DAVIDE at the European Genetics and Anthropology Blog has an interesting post regarding 23andMe’s Ancestry Painting, at “Taking a closer look at your inter-continental ancestry results at 23andMe.â€Â In the post, he describes how to “rummage through the Flash data behind the “Ancestry Painting” presentation†to learn more about the SNPs involved an admixed Ancestry Painting. The post includes the incredibly simple directions:
First of all, you have to make sure you’ve got the free Firebug plug-in installed. Right click on the little bug in the lower-right corner of your browser window, and choose “Enable all panels”. Then left click the same bug icon, which should make a whole new section appear at the bottom of the screen.
Go to the “Ancestry Painting” page, and wait till it loads up your “Chromosome View”. Once it does, select “Response”, and you should get the following link in bold within your new section: POST https://www.23andme.com/you/fetchpaint. Click on it and the desired data should appear.
Hacking My Results:
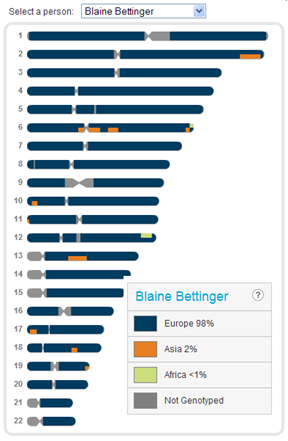
Let’s use the technique to look at my own results (as I’ve mentioned before, I’m not concerned about sharing my results publicly). Here is a snapshot of my Ancestry Painting:
And here are my results using the Firebug plug-in:
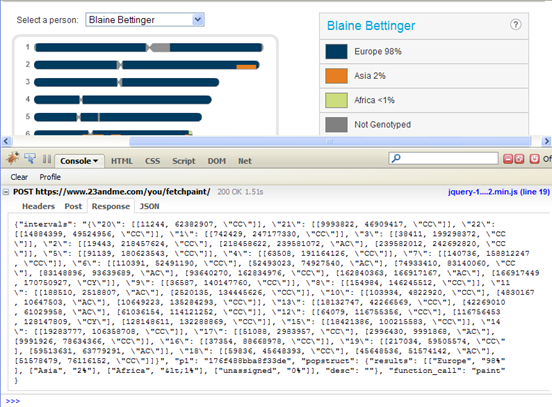
According to the Ancestry Painting (and now the Firebug results), I have Asian segments on chromosomes 2, 6, 10-13, and 17-19, with chromosome 6 being the most admixed. I also have African segments on chromosomes 6 and 12 (In his post, DAVIDE explains why the “Y†stands for African ancestry and the “C†stands for Asian ancestry.):
- 2: [218458622, 239581072, \”AC\”]
- 6: [52493023, 74927540, \”AC\”], [83148896, 93639689, \”AC\”], [162840363, 166917167, \”AC\”], [166917449, 170750927, \”CY\”]
- 10: [4830167, 10647503, \”AC\”]
- 11: [188510, 2518807, \”AC\”]
- 12: [116756453, 128147809, \”CY\”]
- 13: [42269010, 61029958, \”AC\”]
- 17: [2996430, 9991868, \”AC\”]
- 18: [45648536, 51574142, \”AC\”]
- 19: [59513631, 63779291, \”AC\”]
So, for example, my largest Asian segment begins at position 218458622 on one of my chromosome 2 and ends at 239581072.
Yet another interesting tool to use with your 23andMe results.
Great hack! One of our scientists showed me how to do this a while back to identify SNPs on my short European segment (likely from a Spaniard on my mother’s side–she is from the Philippines).
SNPs are a useful tool to quantify LD. The structure of LD along each particular genome or genomic region affects the resolution of association studies [7,8]. For genomes with a slow LD decay with distance, the whole genome may be scanned to identify regions that are associated with a particular phenotype in an association mapping strategy. However, when LD decays rapidly within short distances only nucleotide variation at selected candidate genes may be tested for association with a phenotypic trait