Autosomal DNA is challenging. Although the results of atDNA testing contains vastly more information than either Y-DNA or mtDNA, it is significantly harder to utilize and understand. To wring as much information as possible out of atDNA test, genetic genealogists use every tool available.
One of those tools is triangulation. Although the topic of much debate recently, and subject to several important considerations and limitations, most genealogists agree that triangulation is a valuable tool for working with segments (especially among close relatives).
Unfortunately, there is a lot of misunderstanding when it comes to what is – and what is not – triangulation.
Let’s take a quick look at what triangulation means with regard to atDNA, and why it matters.
The History of Triangulation
The term “triangulation” was not invented by genetic genealogists, although it has been commandeered and now has a very specific meaning in the field. Indeed, the word “triangulation” has been borrowed and its meaning modified in many different fields (see Wikipedia’s disambigulation of triangulation here). That is, after all, how languages grow and evolve, particularly as new technologies are developed.
Triangulation also has a slightly different definition with regard to Y-DNA and mtDNA; the process is very similar, except the ‘triangle’ is often two-dimensional and the third point of the triangle is the MRCA instead of a third test-taker, which significantly lowers the confidence level of the conclusion (see the ISOGG wiki page here).
Why Does it Matter?
The definition of triangulation is important because genetic genealogists who hear or read that a segment has been triangulated assign a very specific meaning to that statement. True triangulation is a very high bar, and thus identifying a segment as being “triangulated” gives it a special status compared to other forms of analysis.
While it is certainly the duty of every reviewer/reader to ensure that a segment is indeed triangulated when it is identified as such, misuse of the term can be misleading and waste the reader’s time when they have to go back and re-analyze the content after realizing that triangulation was not utilized. This can be especially problematic when there isn’t a sufficient amount of evidence to determine whether segments are truly triangulated.
Even worse, using “triangulation” in the wrong context can mislead people who are new to genetic genealogy. Many newbies misunderstand triangulation because the term is so misused, and thus make unsupported conclusions about their data.
So What is Triangulation?
There is only proper definition of triangulation (note that genealogists may differ on the exact words used below, but I posit that the majority of genealogists will agree that this conveys the very specific meaning of triangulation):
- Triangulation – a process or method by which three or more people all sharing an overlapping segment of DNA in common compare their family trees in order to identify a common ancestor or ancestral couple shared by all.
- Triangulation Group (“TG”) – a group of three or more people who all share an overlapping segment of DNA in common with each other. Members of a TG can work together to identify a shared ancestor that might explain the shared overlapping segment of DNA. This is NOT to be confused with an ICW group!
- Triangulated Segment – an overlapping segment of DNA shared in common by three or more people. A segment of DNA can be triangulated even if a common ancestor or ancestral couple has not yet been identified.
Triangulation requires at least three test-takers. The members of a TG must do more than share an overlapping segment of DNA. The members of a TG must do more than be In Common With each other. Triangulation requires that ALL of those test-takers share an overlapping segment of DNA in common with each other.
In Common With (ICW) is NOT Triangulation
In Common With status is often confused with triangulation. While many ICW groups are actually triangulation groups, one must not be confused with the other.
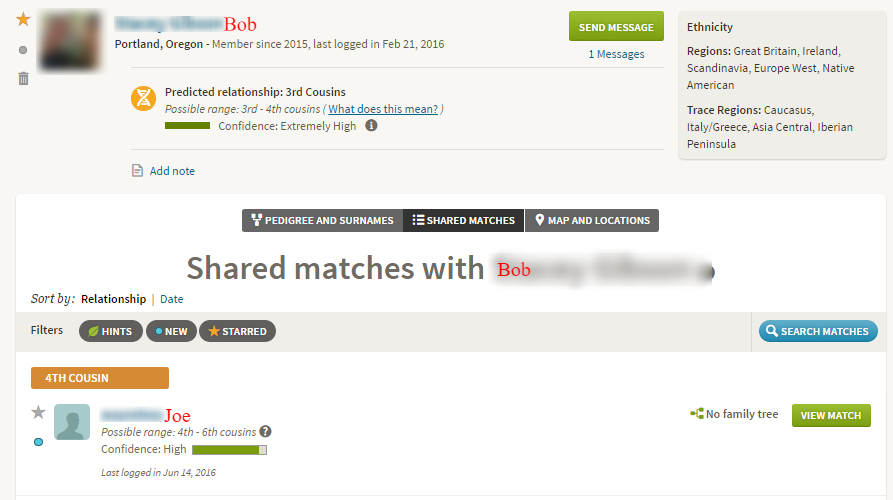
An ICW group means that three or more people all share each other as matches. At AncestryDNA, for example, the “Shared Matches” tool identifies ICW groups. In the image below, Bob, Joe, and I are all in common with each other:
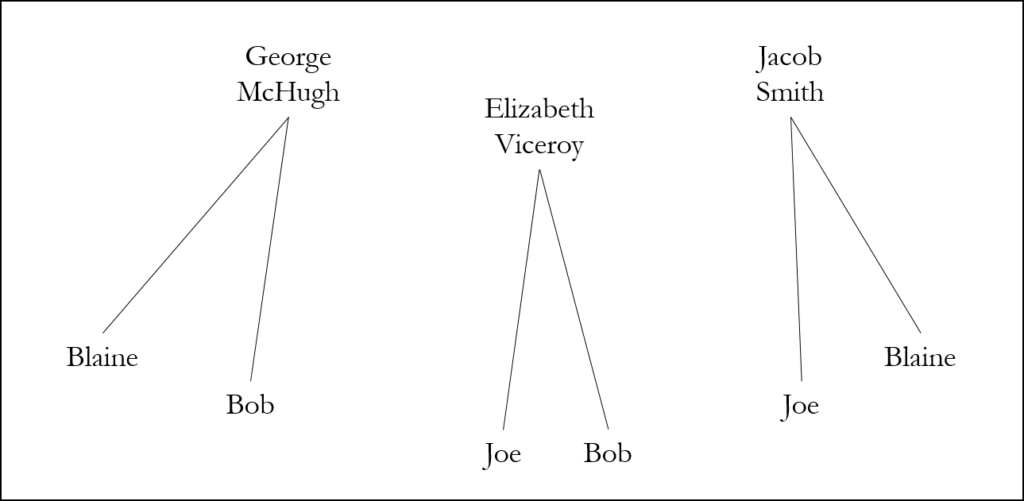
Without segment data, however, we don’t know whether Bob, Joe, and I have one or more segments of DNA in common. It could be, for example, that Bob and I share one ancestor, Joe and I share a different ancestor, and Joe and Bob share yet another ancestor:
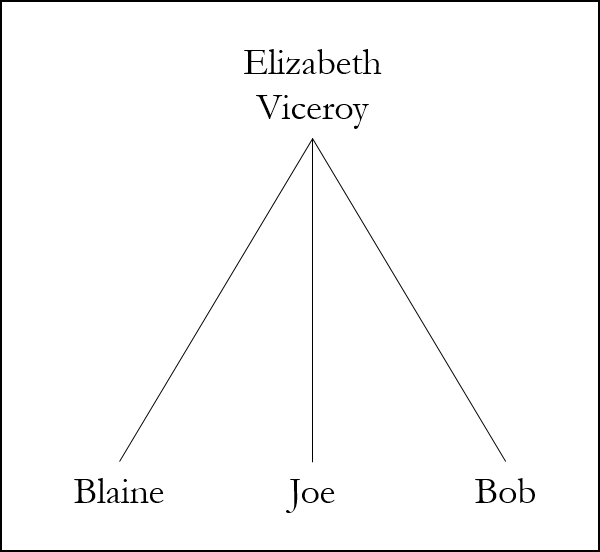
Alternatively, Bob, Joe, and I may share a single common ancestor or ancestral couple in common:
Although there is a wealth of information in Shared Matches, it is not possible to perform triangulation with Shared Matches. It is first necessary to obtain segment information (for example if all members of the group transfer their raw data to GEDmatch), AND then it is necessary to determine whether the same segment of DNA is shared by Joe, Bob, and I.
An Overlapping Segment is NOT Triangulation
On a chromosome browser, some of your matches will have overlapping segments that can look something like the following (a screenshot from Family Tre DNA’s chromosome browser):
 This might be a triangulation group (or multiple triangulation groups), or it might not be a triangulation group. Without additional information about the inter-relatedness of the individuals in this group, we won’t know anything further about these matches. For example, all of these matches might be related to me on my father’s side of the family, they might all be related on my mother’s side of the family, or they might be a mix.
This might be a triangulation group (or multiple triangulation groups), or it might not be a triangulation group. Without additional information about the inter-relatedness of the individuals in this group, we won’t know anything further about these matches. For example, all of these matches might be related to me on my father’s side of the family, they might all be related on my mother’s side of the family, or they might be a mix.
Family Tree DNA offers an ICW tool (also called the Matrix tool) that enables some analysis into the inter-relatedness of the individuals in this group, but even with that information we don’t know if it is a true triangulation group. See, for example, the next section.
ICW and an Overlapping Segment is NOT Triangulation
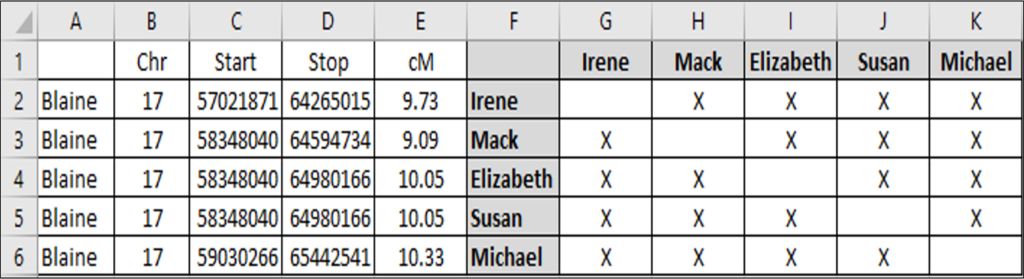
It is also possible to combine segment data and ICW information, such as in the following chart (created using the KWorks tool at DNAGedcom), although it is still not triangulation:
This chart demonstrates that there is an overlapping segment of DNA on chromosome 17 in the region of 57021871 to 65442541. The chart also demonstrates that there is a group of people in this chart that shares DNA (shown by the “x” in the intersecting boxes). For example, everyone in this particular graph forms a single large ICW group:
- Irene, Mack, Elizabeth, Susan, Michael, and Me
Although I share this region of DNA in common with ALL of Irene, Mack, Elizabeth, Susan, and Michael, there is NO guarantee that we all share the same region of DNA in common.
Indeed, there are many different combinations that could explain the ICW graph. For example, the six of us might all share a recent common ancestor that explains the overlapping segment.
However, it is also possible that some of the members of the ICW group share DNA with each other at another location, while still sharing this segment in common with me. They would still all be ICW, but would not all share an overlapping segment of DNA, and would thus not be a true triangulation group.
Many, many people have reported that having an overlapping segment and ICW status results in a true triangulated segment a very large percentage of the time. My own experience agrees with this anecdotal evidence. However, whether they form TGs or not, these ICW groups shouldn’t be called TGs without the necessary confirmation.
True Triangulation
The only way to perform true triangulation is to have segment data and a way to confirm that an overlapping segment is actually shared by two or more genetic matches. This cannot currently be done at AncestryDNA or Family Tree DNA. There are indications of a triangulation tool at 23andMe, but since I am yet to be added to the new interface, I won’t report on that until I am transferred.
Triangulation CAN be done at GEDmatch where it is possible to check whether two or more matches share an overlapping segment of DNA in common (such as using the Tier 1 Triangulation tool, discussed below).
Triangulation can also be done the “old fashioned” way, the way we did it before there were any tools available. A possible triangulation group would be identified using segment data (such as segment data obtained from FTDNA), and then members of the potential group would be emailed to ask whether they shared the identified segment in common with the identified matches. If they responded and said yes, it would be a confirmed or rejected triangulation group.
Triangulation Tools
It is very important to note that tools like KWorks, JWorks, and ADSA at DNAGedcom, and Matching Segment Search at GEDmatch, while incredibly powerful and valuable tools, do NOT perform triangulation. They identify overlapping segments among a group of individuals, and then assign ICW status. Although these ICW groups are often triangulation groups, they cannot be called triangulation groups without knowing whether they all share the segment of DNA in common.
In contrast, the Tier 1 Triangulation tool at GEDmatch DOES perform triangulation. It confirms that all members of a reported triangulation group share the same segment of DNA in common.
Thank you, Blaine!! I’ve been doing triangulation for years now…most people think I’m from another planet!!
Haha! I think most of my ancestors came from another planet! Keep up the good work!
Blaine, that “came from another planet” worked out rather well for Superman!
Haha! Yes, yes it did!
Great post. This will be helpful to share with other cousins who have tested and need a primer on what’s what.
Thank you Michael! I’m so glad you found it useful. The many, many intricacies of triangulation that I didn’t get to cover here are important, but this blog post is a good primer on the definition of triangulation.
I feel like you wrote this specifically for me! Thank you for clearing this all up! Your writing is both informative and accessible.
Thank you Rachel, that’s so nice of you to say!
I use the one-to-one tool to check each member against the others in the hypothesized triangulation group. I just thought that ought to be said since using one-to-one doesn’t require having the Tier 1 tools (as much as they are deserving of the financial support).
Absolutely, thank you for highlighting that option Patti! Truly one of the things that makes GEDmatch so powerful for genetic genealogists.
I might just add that the three people should have independent routes to the MRCA. If two of them are siblings, or parent/child, they are essentially saying the same thing twice.
Terrific addition, Ann!
Technically isn’t your statement also true for two first cousins who further on match with a third cousin?
Beginner here — good article but need clarification on Ann’s and this point. It sounds as though the anwer is yes–that triangulation to a common ancestor is not possible if two distant cousins get there through the same child of MRCA and the third distant cousin gets there through a different child of MRCA. Until now I thought I could rely on Gedmatch triangulations and just find the MRCA in a triad.
I was wondering about that, thanks for clarifying.
You’ve done a great job explaining triangulation concepts and giving due warning of pitfalls and limitations. By the way, I really like the title of the blog :).
Haha, thank you!
I’m very thankful that you took the time to write this as it is very clear and easy to understand. It is the first time I have actually understood what the various terms mean altogether! Good job!
Wonderful Jim, that’s such good news. I spent way too long on this post, so it’s nice to know it was well worth the time.
Blaine, I want to second what everyone else has said. Very helpful to me too, I’m glad you took the time to make this so clear.
Blaine, Some time ago I placed a rough and ready old fashioned way, using excel and lots of cousins on a couple of Facebook forums to identify by example the “Triangulation” method I use. Within minutes, back came criticism that I had not taken any notice of the “In Common With” matches. It did not seem to matter to those writers that I had proven my method by fact of known family (some 6 cousins all at various paternal ancestral positions ranging from 1st to 6th) plus my siblings.
And now you put into words so nicely exactly my same criteria that I used. I am feeling on top of the world! (I had long given up on using “In Common With”, except in occasional curious moments when I am puzzled by what appears at first glance to be an overlap, but later turns out to be sheer coincidence). THANK YOU!
Gail, even though in-common-with doesn’t tell you where testers are matching (and if they are matching on the same segment), it can give you other people who are possibly related in the same way which may result in another triangulated group for the same ancestral couple. I did that just tonight by using people who matched two kits, and found not only all the people triangulating on the one segment of interest, but also another segment shared by three, and several others shared by two. In the past, I have gotten those segments to triangulate when more people were tested later.
That’s great Gail! Happy to be of assistance!
Since this will be read by beginners, you need to clarify the sentence “Triangulation requires at least three test-takers.” If two of the three are parent-child, grandparent-child or siblings, you cannot count them separately.
Ooops. I see that Ann said the same thing.
Thanks for adding Israel, that is definitely an important point!
A great article, Blaine. It’s good to separate TG and ICW.
I often use the logic that all shared segments (between me and each Match) are either IBD (from a Common Ancestor) or not IBD. All IBD segments are maternal or paternal – the only two choices. So if a shared segment does not triangulate with a TG on either side it must not be IBD. And if we use large segments we are sure are IBD (say over 15cM), then if they are ICW segments in a maternal TG, and not ICW with segments in a paternal TG, we are pretty close to certainty that those segments belong with the appropriate TG. I color code Triangulated segments red and blue when I know which side. For segments which I determine are aligned with one TG or the other (using ICW, ethnicity, same email, etc), I color code them pink or light blue to indicate that they haven’t passed the rigerous TG test, yet. But I always go back to the original concept that all shared segments have to be in one of three categories: maternal, paternal or not IBD (a false segment). You make a very good point that an ICW list or matrix forms cannot, alone, categorize the overlapping segments. Bravo for a great post.
Thanks Jim, that’s high praise coming from you! Your point about IBD or not IBD is very well taken, and it is exactly the reason why the table above only has one ICW group! As soon as you have two ICW clusters at a single segment, the possibilities narrow significantly.
Your color coding is an excellent example of proceeding with what you have while also keeping a note of the ICW and TG status. Thank you!
Thanks! Slowly, but surely, I will understand all of this. You just helped me get closer to that mark!
Renate
Wonderful Renate, so glad this helped!
So how important is it that three individuals share a segment in common and what use is it?
Joe – that is a million dollar question! My only goal with this blog post was to ensure that everyone understood the definition of triangulation compared to other analyses. How to use triangulation, including its benefits and limitations, is much more complicated. If recommend the “triangulation” page on the ISOGG wiki as a starting point.
Just want to say that I appreciate your blog. I may not always understand, but your clear explanations go a long to help! Thanks, John
Thank you John!
When I work with a TG, I am more comfortable working with 4 people, instead of 3. Yet, I did not have a name for that, since tri means 3. Seems you call more than 3 a “multiple triangulated group.” That is certainly better than Quadangulated. Yes, I know that is not a word. LOL
Absolutely, a TG can include just 3 people, or 100 people!
Wonderful blog post Blaine! I do think people misunderstand triangulation and how it differs from Y and mt to atDNA. ALSO, there is much confusion out there about ICW which always needs clarification among those new at genetic genealogy.
Thank you for all your wonderful work!
Thank you Emily! And it sure was great seeing you California!
Dear Blaine,
Maybe it would be more clear to others if you also used the “Identical by Descent” or “Identity by Descent” which is ideally what your triangulation also seeks. ICW looks to me more like having a common haplotype.
George Cicila
P.S. why are geneticists sticking with centiMorgans (cM) when measuring the amount of shared chromosomal material among distant relatives (third cousins and up). Wouldn’t mega basepairs more accurately reflect the amount of genetic material in common?
Great points, although IBD is a bit more advanced than I wanted to be for this blog post. ICW simply means that two people share at least one segment of DNA above a company threshold. It might be IBD, but then again it might not.
Among other reasons, using centimorgans instead of base pairs actually gives us more information about a segment. If measured just in bp, we don’t have any information about the probability of recombination for that segment. If measured in cM, we get that additional information about the probability of recombination.
Would you write about cms vs base pairs and recombination…PLEASE!
Thank you Blaine. We genetic genealogy newbies need more posts like this one: plain English explanations of concepts beyond the basics. I have done enough reading, listened to enough webinars, and attended enough sessions at conferences to know that there are too few people providing “how to” advice regarding what to do with DNA test results.
That is so nice of you to say, I greatly appreciate it! I agree, we need a lot more education in this field at all levels.
I do triangulation at 23andMe using the Family Inheritance: Advanced feature under Ancestry Tools. If I match A and B at a segment, I then put A into the Compare slot and see if A matches B at that segment. I think this is a valid process to confirm a shared ancestor.
Thanks very much for this clear & concise explanation of Triangulation vs ICW “In Common With”! I will be sharing it with others in my genealogy group who are struggling to understand autosomal DNA analysis!
Wonderful Betty, please feel free to share with your group!
You’ve provided a very clear and necessary explanation of what true triangulation is. A few days ago, I coined the terms: Single Match Triangulation (SMT) and Double Match Triangulation (DMT) to represent the use of a single person’s matching segments, versus using two persons’ segments: http://www.beholdgenealogy.com/blog/?p=1767
Double Match Triangulation, as you explain, is necessary for true triangulation.
Note the follow-up I added at the end of my post making reference to your article posted here.
Great, thank you for the link Louis! I’m looking forward to studying your blog post.
I read your article on triangulation as it applies to DNA. I use what is “old school” methods without DNA(don’t understand DNA that well) eg: land titles, death certificates, census, grave stones, shipping manifestos and wills. I have been Y-DNA tested(I have taken all the Y-DNA tests available) and placed in a Group of early North Carolina Phillips family members. Of course, the y-DNA has helped identify many living relatives, the ancient ones have no DNA to prove who they are so other means must be used. I am a member of FTDNA and the first thing they did was assign a volunteer genealogist to assist me in finding the correct relatives. I have corresponded with many cousins who I never met and some of these cousins are versed in DNA and many also have family trees that are shared with the Group. Thanks for writing about DNA and triangulation. I am sure other Phillips members who are versed in DNA will appreciate al your efforts. Jerry D. Phillips
Jerry – DNA won’t work without all the traditional genealogy research! They always work best together.
I appreciate your efforts to bring clarity to this issue. However, I’ve never understood where the notion came from that it’s necessary to have three people to triangulate. The analogy with Y-DNA seems to work perfectly well with autosomal DNA because we are using two known data points (eg, two third or fourth cousins with well documented trees) to assign a segment to a specific ancestor. Perhaps adding a second or a third cousin to the mix would provide additional confirmation but I don’t see why it should be deemed necessary.
Yes, Leah and I discussed this issue in the ISOGG Facebook group comment thread. Identifying a MRCA certainly does not require three genetic matches (although the confidence level that the correct ancestor is identified is very low with just two people, much better with three, and even better with more than three).
However, the real reason triangulation requires three genetic matches is because the community has adapted the term to mean that. If you tell a genetic genealogist that a segment is triangulated, they’ll believe that you utilized the DNA of three or more genetic matches.
The confidence level is only increased if you have additional close relatives testing to confirm the triangulation (ie, first, second and third cousins).
My concern is that I see people who are claiming that they’ve triangulated a tiny segment between three or more distant cousins and they are then claiming this as proof that they all share descent from the same ancestral couple. They ignore all the caveats about the importance of phasing and chromosome mapping, and fail to to consider all the more plausible reasons which would explain why several people would match on the same small segment (ie, excess IBD, descent from multiple ancestral pathways).
You’re absolutely correct, the field definitely needs to do a lot more work exploring and testing triangulation. To do so, we all need to use the same definition of triangulation, which was one of the reasons I wrote this post.
For large segments (i.e., high likelihood of IBD), the segment can be triangulated even if no MRCA has been identified. In fact, this is the current status of many or most of my large shared segments.
It might be interesting to do a survey to find out more about the size and nature of triangulated segments by population. I don’t have any large triangulated segments. All my apparent triangulated segments are on small segments under 15 cMs, and mostly under 10 cMs. I did start to investigate these but concluded it was a complete waste of time, and my efforts were better spent looking at larger segments rather than triangulated segments. I’ve never bothered to check if these are “true” triangulated segments. It didn’t seem worth the effort.
Not if it is YDNA, where the term was first used to denote two present-day people and one in the past: the MRCA. That term was derived from surveying and in that use it is a direct parallel and a good metaphor.
Autosomal “triangulation” discussions often focus on how many people are at the base. It can be good to consider things pairwise towards an ancestor. Further connection between present-day people often only works when we understand the relationship of each pair to an MRCA. Then we can move on to fitting them together, if possible.
I understand the concept – and would like to do it. However, I come from an endogamous population. About 18 months ago I bought the Tier 1 utilities and was very disappointed that it was unable to run my data (Just as DNAGEDcom blew up). I have tested 20 known cousins – and would like to be able to triangulate them with 30 or 70 or a relatively small number of unknown matches at the same time. I would be very happy to triangulate in batches – provided that the batches were larger than the 5 that I can run in FTDNA’s chromosome browser. Any idea when I will be able to use some of these neat utilities?
It’s definitely going to be a challenge with an endogamous population. Have you tried KWorks at DNAGedcom? I just used it yesterday, so I know it is up and running. That will create groupings where there are overlapping segments and ICW segments, which is very close to triangulation. And since you have close cousins tested (presumably on both sides of the family), you’ll be able to form very strong TG groups (see Jim Bartlett’s comment above, for example).
Blaine,
This is a much needed post. It certainly addresses and clears up some of the confusion about what constitutes true triangulation. Thanks for posting this.
I enjoy your blog and I keep my email subscription to it because I know you aren’t going to fill my inbox with frequent posts, but when you do post it is ALWAYS worth reading.
Great blog!
Janis
What a very nice thing to say, thank you Janis! It means a lot to me that people take a few minutes out of their day to leave a comment or feedback to let me know what they like or don’t like. Thank you!
Blaine,
Our discussion with you on this topic last week in the IGHR Genetic Genealogy class showed that even those who have been working in/learning about the field for some time can benefit from more clarity on this concept. Thanks for a great follow-up! “D1” and I will certainly share it with our SIG here in Louisville.
Thank you Debbie! It sure was great seeing you and D1 last week!
This is an awesome account I have forwarded it to every person I know who has done genetic testing. Thank You.
These discussions are so helpful. When a maternal female cousin and I had our mtDNA and autosomal tests done by FTDNA we were each other’s closest matches. A third person was later shown to be an equally close match to me (without leaving any contact details) I worked out that it had to be from my paternal side as this third person did not overlap our mtDNA results. The term triangulation seemed to cover the process when I heard of it later. I have tried the Matrix process with the 5 entries allowed. Next step to download the raw data I expect. Ancestry results now in.
Outstanding pedagogy! I wish this lesson had been available a year ago when I was struggling up the humongous (for this octogenarian, at least) learning curve of genetic genealogy and tripped over this gosh-awful term “triangulation,” which like most newbies, I suspect, I had conceptualized in the surveying (and Y-DNA) sense as simply locating an ancestor at the intersection of two lines of descent. To me — again, I bet I speak for most of the unanointed — the term neither says what it means nor means what it says, and serves only to obfuscate an already difficult concept.
Somewhat like a term I spent my career as a professor of medicine trying unsuccessfully to remove from medical jargon, “blood thinner,” which is foisted off on their patients by doctors and nurses who must think they are unable to comprehend the big word “anticoagulant,” which does not affect blood viscosity or blood flow. Patients actually occasionally self-regulate the dose of their “blood thinner” because they feel sluggish (blood too thick) or cold (too thin), causing bleeding or clotting problems. All the more unfortunate in this example because there is an easily understood alternative term, “anti-clotter” (with two fewer letters and only one more syllable) which does say what it means and means what it says.
Sadly, “blood thinner” and, I fear, genetic genealogical autosomal “triangulation,” both fall in the category of Entrenched Errors, so thoroughly established in the jargon that they cannot be corrected. And, after starting to write an article (because the best way to learn a difficult subject is to teach it) which I hoped would be as comprehensible as yours, to be entitled something like “____________ for Genetic Genealogy Dummies,” I gave up trying to find a suitable substitute for that seductively simple, even melodious, term “triangulation.” Nevertheless, I hope this comment will motivate a search for a better way to encapsulate the true meaning of the process you describe so clearly and well. (OBTW, I like your title much better than mine, too.)
Addendum: In Blaine’s outstanding book, “Family Tree Guide to DNA Testing and Genetic Genealogy,” he has used (?coined) the terms “Tree Triangulation” and “Segment Triangulation” to deal with this issue. Will others adopt that terminology?
I got a question. I have a relative on gedmatch who apparently shares enough autosomal DNA to be considered not just a cousin but a full sibling. Unless my parents adopted out some unknown sibling of mine, I possess one full sibling & one female half sibling [the relative is male apparently]. 3523 versus the 2678 I get with my sibling, 1.0 generations versus 1.2 generations.
Thing is, is this relative doesn’t share any chromosomes with myself or my sibling. Nor for that matter grabbing a bunch of random relatives and using gedmatch’s chromosome segment matrix anyone else.
So is he a relative or some glitch?
Hi Blaine,
Your blog is terrific. It seems to me that one ought to be able to take autosomal DNA results from a bunch of first cousins and stitch them together to make composite autosomal DNA result of their common grandparents.
To be clear one wouldn’t be able to differentiate between which grandparent contributed which DNA but with enough first cousins one should be able to get an approximation of what the sets of autosomal DNA were for the grandparents.
Admittedly this would not be triangulation, but it could be incredibly useful. Are there any groups developing tools to do this kind of thing?
Let’s say I have three kits and I want to see if they form a triangulation group. How can this be done using GEDMatch? The multiple kit analysis tool allows me to enter three or more people, matches A to B, B to C, etc, but as I understand your blog, if A-B match, B-C match, B-A match and all three groups overlap on a segment they do not constitute a triangulation group but rather an ICW group.
You’re right, without that extra piece of information, that is only an ICW group.
But, you can just do a one-to-one comparison of the other individuals to each other to see if they share that segment in common.
I’m confused. It sounds like Mike had compared A to B, B to C, and C to A and found a segment that they all overlapped on. Then it sounds like you said that wasn’t triangulation, but you could do a one-to-one comparison of the “other individuals” . . .
What other individuals?
Carol, I was confused, too. But, my confusion was with Mike’s question. Your comment didn’t properly repeat Mike’s question. He compared A-B, B-C, and B-A (repeating the first test, rather than what you asked about, and what Mike should have done if truly trying to triangulate, the C-A comparison).
Blaine appears to me to have addressed the comment the way it was written and suggested using the one-to-one test between each pair of members of the test group – ie. the missing C-A test.
This blog post is essential reading !!!
I only wish I had read it earlier.
I have often seen the term ICW, and understood that it means in common with, but this post is the only time that I’ve seen context given to the term.
It also gives an explanation to the difference between ICW and TG.
Great article, Blaine.
One thing I don’t recall seeing and may be an erroneous assumption on my part is that the lines from each member of a TG to the MRCA must merge only at the MRCA. Is that correct?
Thank you,
John
Okay I am going to jump right in.
Using my mother’s kit I have a second cousin on her mother’s side and a fourth cousin on her father’s side, I discovered yesterday that they overlapped on the same segment on one chromosome but they weren’t matches to each other.
Someone pointed out that this was because we inherit two of each chromosome so one would match on the paternal and one on the maternal and that was why people talk about triangulated matches. That got me interested to learn more and led me to here.
So what does the Tier 1 triangulation on GEDMATCH actually do that can’t be done via one to one matching and chromosome browser?
Also am I right in thinking this only works well if one person is a close relative? I have people who match each other and me but not on the same chromosome yet I know we are all related to each other, so I presume we have just inherited different segments from our mutual ancestor.
Thanks for making this a little clearer though.
Question: I have two cousins that triangulate (GEDMatch one-to-one comparisons with a potential cousin (all separate pairs match on same segment.) The known cousins are my 5th-6thcousins, the potential cousin would be our eighth cousin. Ditto on another segment with two different known cousins and the same potential 8th cousin. Due to the large degree separation to the potential 8th cousin, the SNPs are ~200 and at 3, and 4.2 cM on the two different triangulated segments. What is the confidence level that we share common ancestor? I’d suspect it’s a shoe-in. The segments are not on Chr 15.
Blaine (and all the nice people who read this):
Why do you think FTDNA and Ancestry do not give us a true triangulation tool?
Why do you think DNAGEDCOM does not give us a true triangulation tool?
I am really tired of working on triangulation because it simply takes too much time.
Do the atDNA testing companies not want us to more easily discover our common ancestors?
Thanks
Hi Blaine. This is the most informed I.ve gotten in regards to matches through DNA. I appreciate how you explain the difference of being ICW and TG. I will read through this several more time as well as the response. I have question which may also be commented on but I.m wondering if each chromosome represents a certain trait of if certain chromosomes indicate whether paternal or maternal. Thanks again for dull your time and research. Grrard
Thanks for this informational article, Blaine! As a Genetic Genealogy novice, I was wondering about your statement that the only Triangulation tool that performs this type of testing is the Tier 1 Triangulation tool found at GEDmatch.
Since it’s been over 3 years since you wrote this article, I wondered if anything has changed? Are more companies testing for this type of Triangulation or has there been any update on your article?
I read another article that claimed that while this is the Gold Standard, it’s rare to find several people with shared segments, due to random recombination.
I’m interested in your thoughts about these two points.
Thanks!
Blaine this was a very helpful article. Could You comment on how many people typically can share triangulated segments? I just watched a lecture by Maurice Gleeson where he said that 5 people sharing a triangulated segment is very rare and six was almost unheard of. I find this puzzling as I have maybe ten or more on MyHeritage that are triangulated from a ethnicity I know is in my family although the start and end points aren’t exact but a number are close. Thank-You