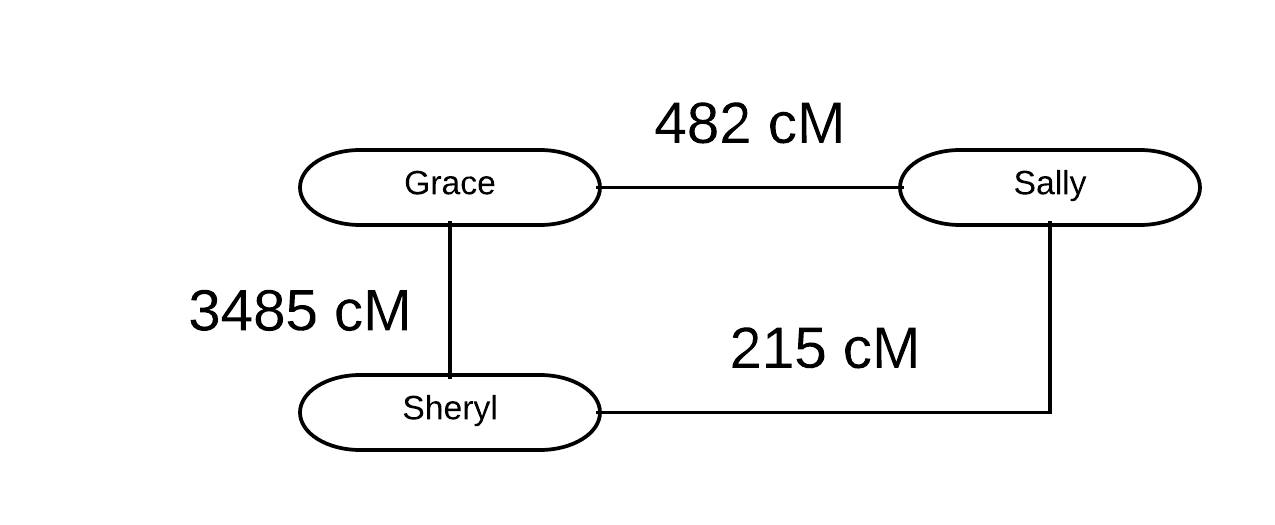
Have you experienced this? You’ve identified a very clear cluster that includes numerous DNA matches that all descend from a single family, but you have no idea how this family links into your family tree. Try as you might, and despite building numerous trees, you can’t seem to figure out how these DNA matches and this single ancestral family link into your family tree. If this sounds familiar, you have an Unlinked Family Cluster!
Defining “Unlinked Family Cluster”
An Unlinked Family Cluster is a very specific phenomenon in genetic genealogy, one that is becoming increasingly common. We see more and more of these clusters for various reasons; the matching databases get larger and larger meaning that these clusters get larger and easier to identify. Additionally, the more we work with our closest DNA matches, the more we have very promising “left behind” matches that don’t fit into our known ancestry. Often, these “left behind” matches form a shared match cluster or a triangulated cluster around a specific family. This is an Unlinked Family Cluster.