Five-Part Series on Visual Phasing:
- Part I – Explaining visual phasing and identifying/labeling recombination points (November 21, 2016)
- Part II – Assigning segments of DNA (November 22, 2016)
- Part III – Using cousin matches to identify which grandparent provided the segments
- Part IV – Mapping my own chromosome using the visually phased paternal chromosomes
- Part V – Using the mapped DNA with new matches
This weekend, I spoke at a meeting of the New England chapter of the Association of Professional Genealogists, and it was a wonderful group. One of my talks was about “Chromosome Mapping.” Unfortunately, since the talk was only an hour, we didn’t have time to discuss “Visual Phasing,” a chromosome mapping methodology. Instead, I promised to finish this blog post to explain the process. As I was writing, the blog post turned into a 5-part series!
Quick Summary
- What is it? A method to assign segments of DNA to the test-taker’s four grandparents.
- Why use it? To identify which grandparent gave the test-taker which segments of DNA (eliminating 75% of the family tree to search for MRCA).
- What do you need? Autosomal DNA of three siblings uploaded to GEDmatch.
Visual Phasing
Visual Phasing is a process by which the DNA of three siblings is assigned to each of their four grandparents using identified recombination points, without requiring the testing of either the parents or grandparents. Although the process does not automatically reveal which segment belongs to which of the four grandparents, matching with cousins provides this identification as a further step of the process.
Kathy Johnston developed this process some time ago, and first posted about it in the Family Tree DNA Forum. As shown in the figure below, there are two PDF documents available for download to explain the method with both images and text. However, note that you must be a registered member of the forum (free) to download the documents.
My understanding is that Randy Whited also independently developed this process. I attended his excellent lecture on visual phasing at SCGS Jamboree in June 2016, and an audio recording of that talk is available for purchase here (Session# TH 023 entitled “Reconstructing Grandparent DNA Using Sibling Results” for $11.00).
Visual Phasing is an incredibly valuable tool. Although requiring three siblings creates a considerable barrier for many people, it can be extremely valuable for genetic genealogists interested in chromosome mapping. For example, as we’ll see, I have an adopted great-grandmother, and using visual phasing I can identify entire portions of my chromosomes that came from her, which could prove to be beneficial to my search.
NOTE: although visual phasing can potentially be performed with just two siblings and close cousin(s), it is considerably more challenging. I strongly recommend starting out with three siblings (either your own family or someone else’s family).
Other Resources
In addition to Kathy’s PDF documents and Randy’s recording, there are several other resources. Joel Hartley has been publishing the results of his visual phasing (see “My Big Fat Chromosome 20”), as has Ann Raymont (“Chromosome Mapping with siblings – part 1” and “Chromosome Mapping with siblings – part 2”). Ann’s blog posts contain a lot of details about the whys and hows of visual phasing.
What You Need
You need three siblings who have done autosomal DNA testing and transferred their results to GEDmatch. The testing company doesn’t really matter, even if you’ve tested all three at three different companies. Trust me!
I perform visual phasing in PowerPoint because it gives me a great deal of freedom to manipulate screenshots and add annotations, but it isn’t perfect. I’d love for this process to be semi-automated, at least creating an output comparison for Step #1, below.
STEP 1 – Setting Up
Visual Phasing works by identifying recombination points in the DNA of the three siblings. As will become clear, a recombination event in one sibling will affect how she shares DNA with the other two siblings.
Accordingly, the first step in visual phasing is to compare the DNA of the three siblings to each other in GEDmatch using the One-to-One tool. We’re going to work on one chromosome at a time, and I recommend starting with the X chromosome (especially if one of the siblings is male, since he’ll only have one X chromosome) or one of the shorter chromosomes such as 20 through 22.
Capture a screenshot of the comparisons, and paste them into PowerPoint.
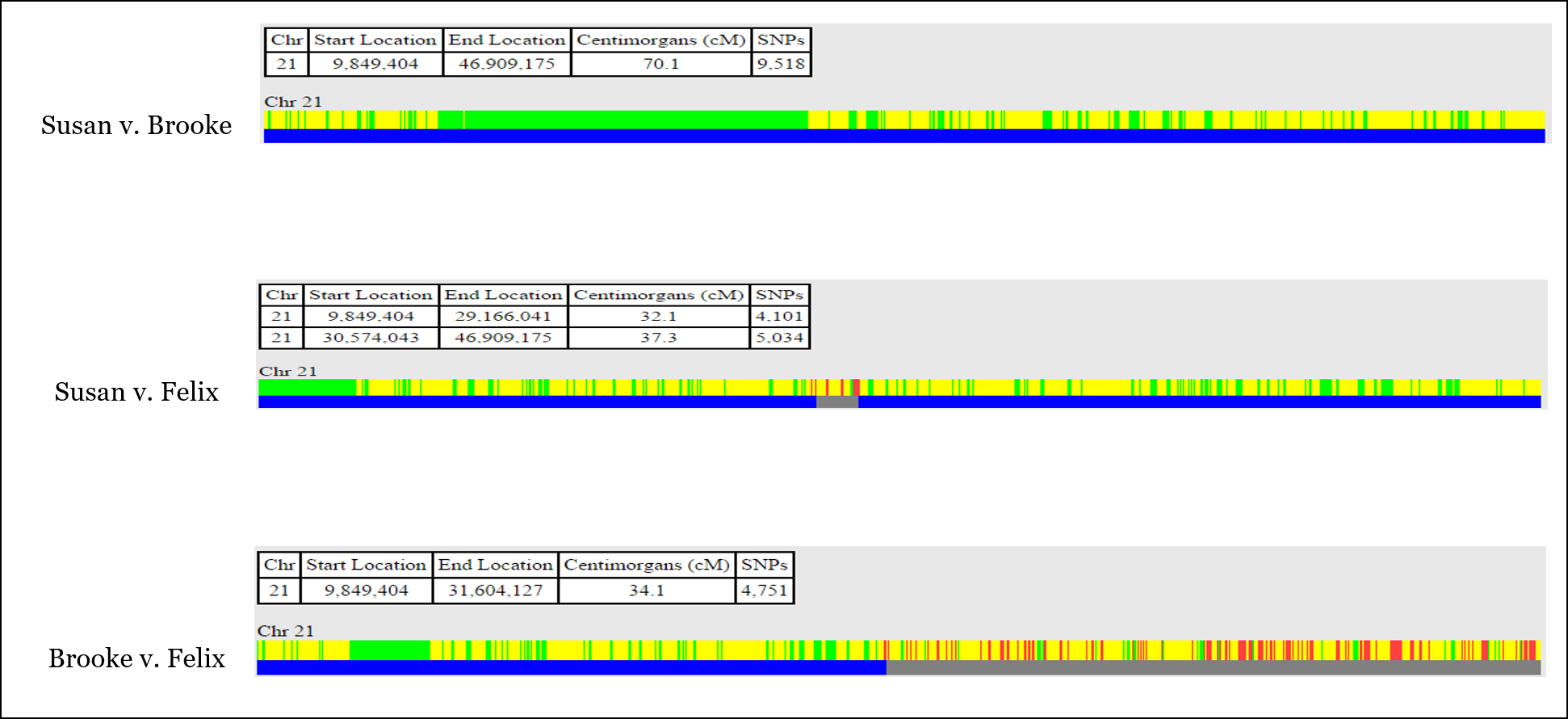
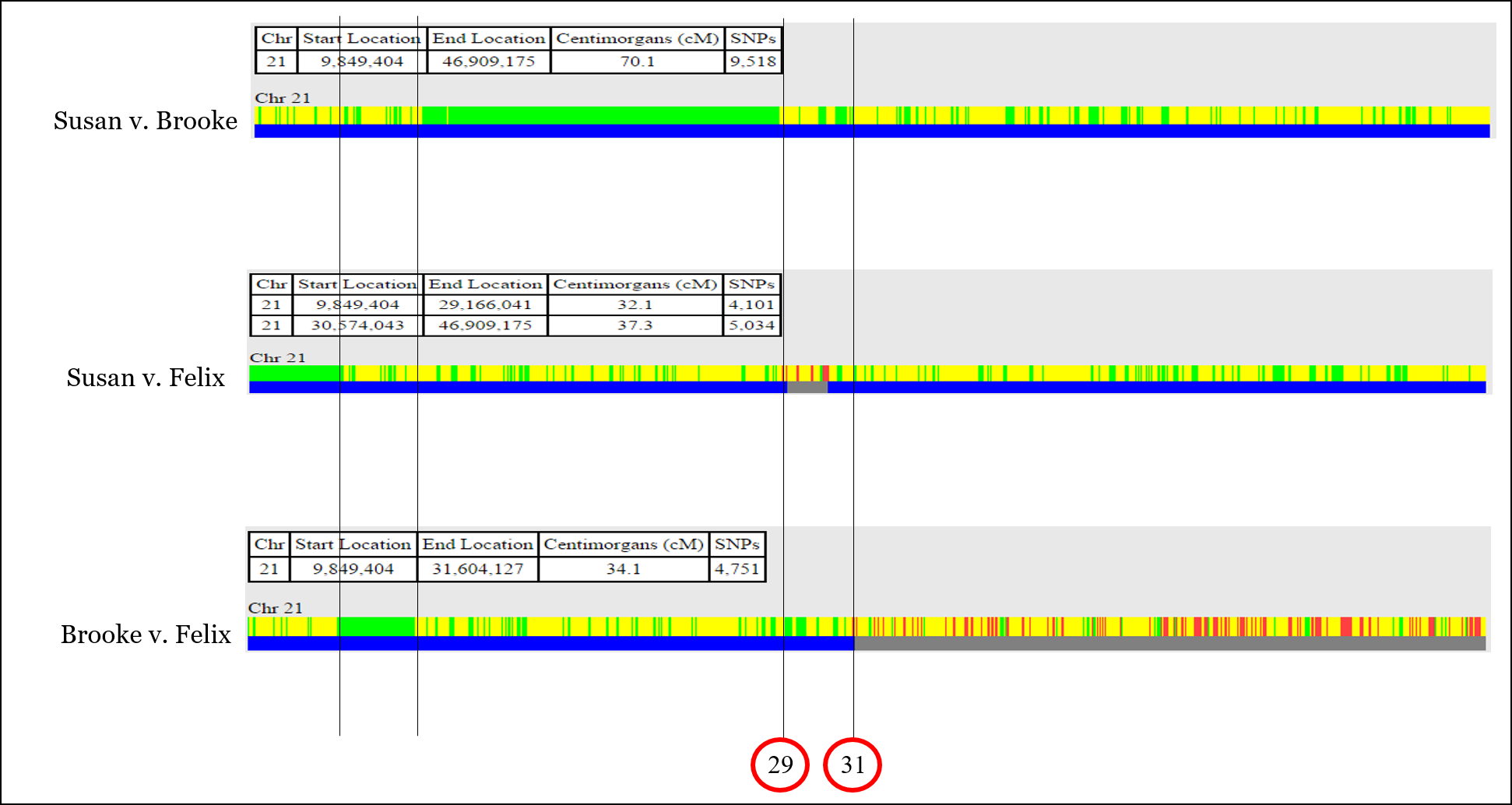
In this example, we’re going to be looking at Chromosome 21 in three siblings, Brooke, Felix, and Susan:
With this information, you can identify most or all of the recombination that took place when the sperm and egg were created for each of the three siblings.
In One-to-One comparison, you’ll usually see both half-identical (yellow) and fully-identical (green) sharing (but not on every chromosome). Remember that we’re actually comparing TWO CHROMOSOMES of each person at each and every point, so sometimes full siblings will share DNA on only one of their chromosome pair, while they will also share DNA on both copies of their chromosome pair.
STEP 2 – Identify and Label the Recombination Points
Now we can identify and label the recombination points. Here is the first key point:
KEY POINT #1 – Anywhere there is a change in the sharing status between two siblings, there must be a recombination event in at least ONE of the siblings (and sometimes both!).
For example, a switch from sharing a yellow segment to sharing no DNA means there was recombination at that point in one or both siblings. A switch from a yellow segment to a full segment means there was recombination at that point in one or both siblings. And so on.
In the following figure, each of the recombination points (i.e., each change in sharing) is identified by an arrow:
Now, in PowerPoint, draw a long vertical line through each recombination point. Each line should intersect at least two recombination points:
Often, this is where you’re going to run into the first problem, namely that the line doesn’t always seem to intersect at least two recombination points. This happens for a variety of reasons. Most commonly, the chromosome visualizations don’t always line up perfectly. Second, sometimes the start and stop locations are fuzzy (Ann Raymont mentions this in her terrific blog post here: “Chromosome Mapping with siblings – part 2”). Third, sometimes there are recombination events in two siblings at one place, which can cause some difficulty.
KEY POINT #2 – Do NOT get too stuck on recombination points. Trust me, getting frustrated with recombination points that don’t line up can quickly derail a phasing project! “Close enough” is just fine when trying out the first few chromosomes.
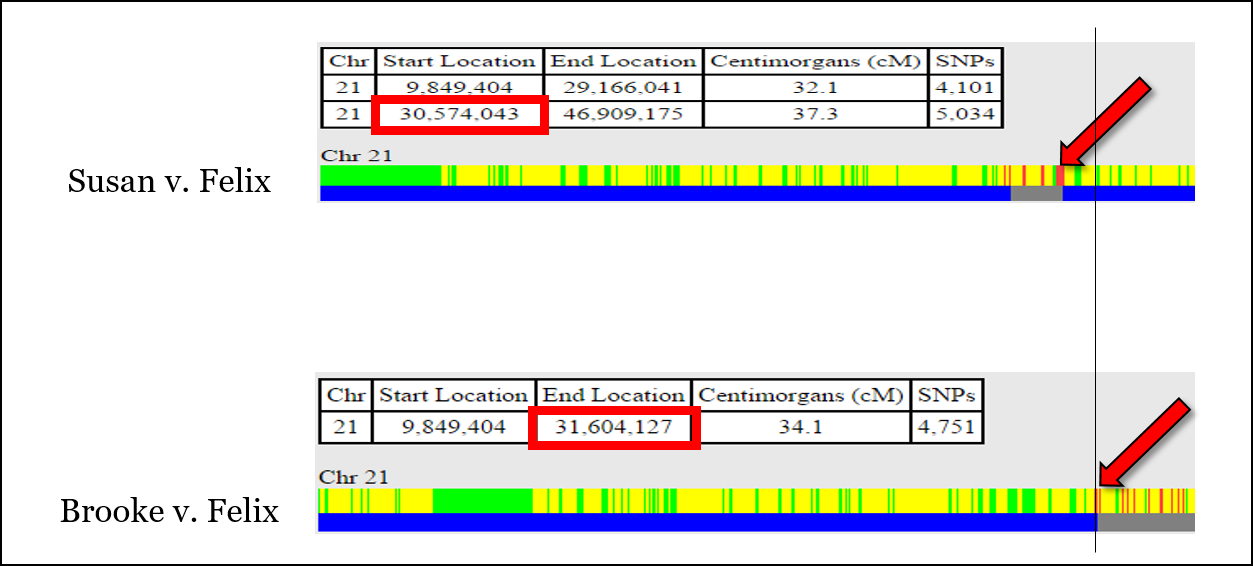
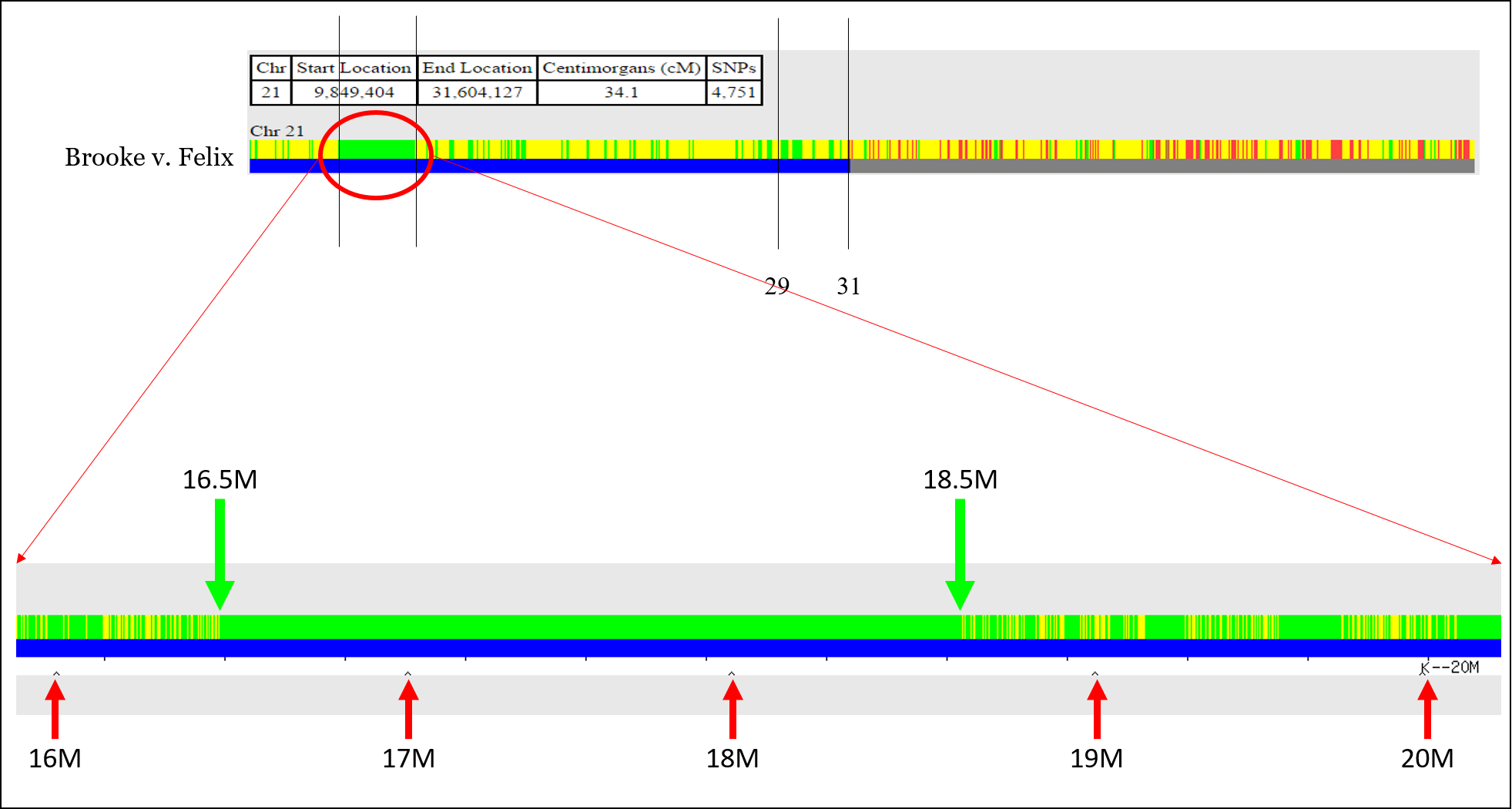
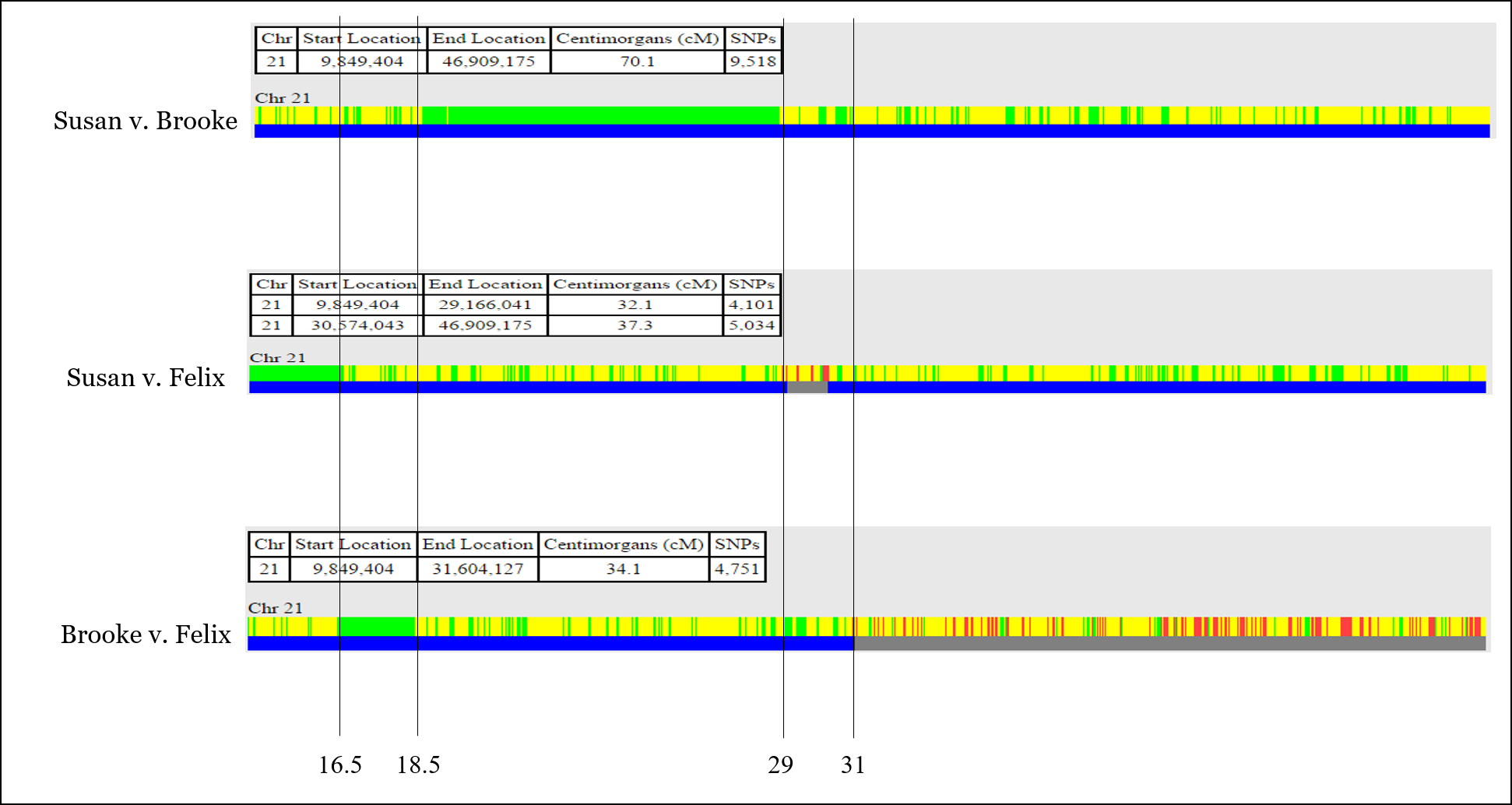
For example, there is an issue aligning the recombination event shown below, which is at position 30,574,043 according to the comparison of Susan v. Felix, but at position 31,604,127 according to the comparison of Brooke v. Felix. This is unlikely to be “fuzziness.”
It’s very easy at this point to throw your hands up and jump to another chromosome. But for now, we’ll put a recombination point around 31,000,000 or so.
I also like to label the recombination points with a number for easy reference. The start and stop positions for each yellow segment (sharing on one chromosome) is provided by GEDmatch.
However, you’ll need to take an extra step to get the start and stop point for green segments (sharing on both chromosomes). This is a great trick that I just learned recently (via Sue Griffith at “Obtaining FIR Boundaries on GEDmatch using the Little Tick Marks”) is to perform a One-to-One comparison, but to click “Full resolution.”
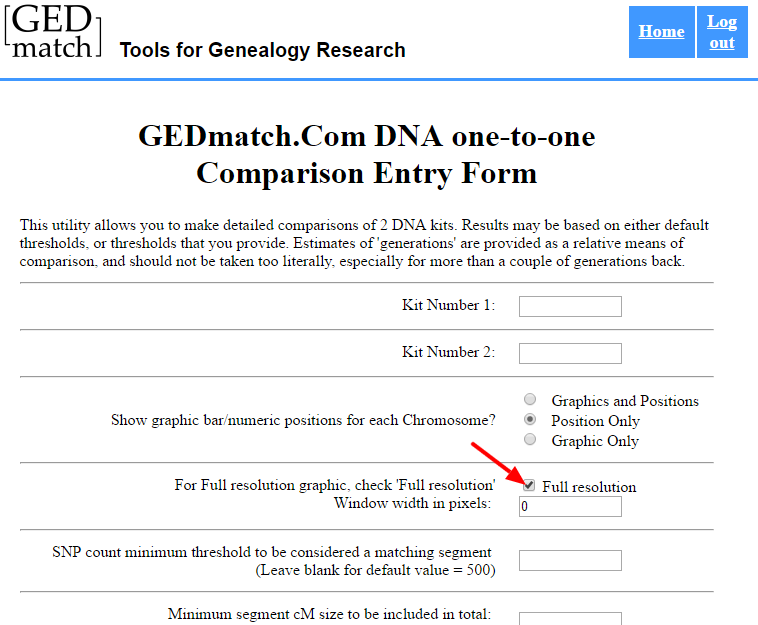
This will expand the chromosome and show megabase positions, and you can obtain the start or stop position:
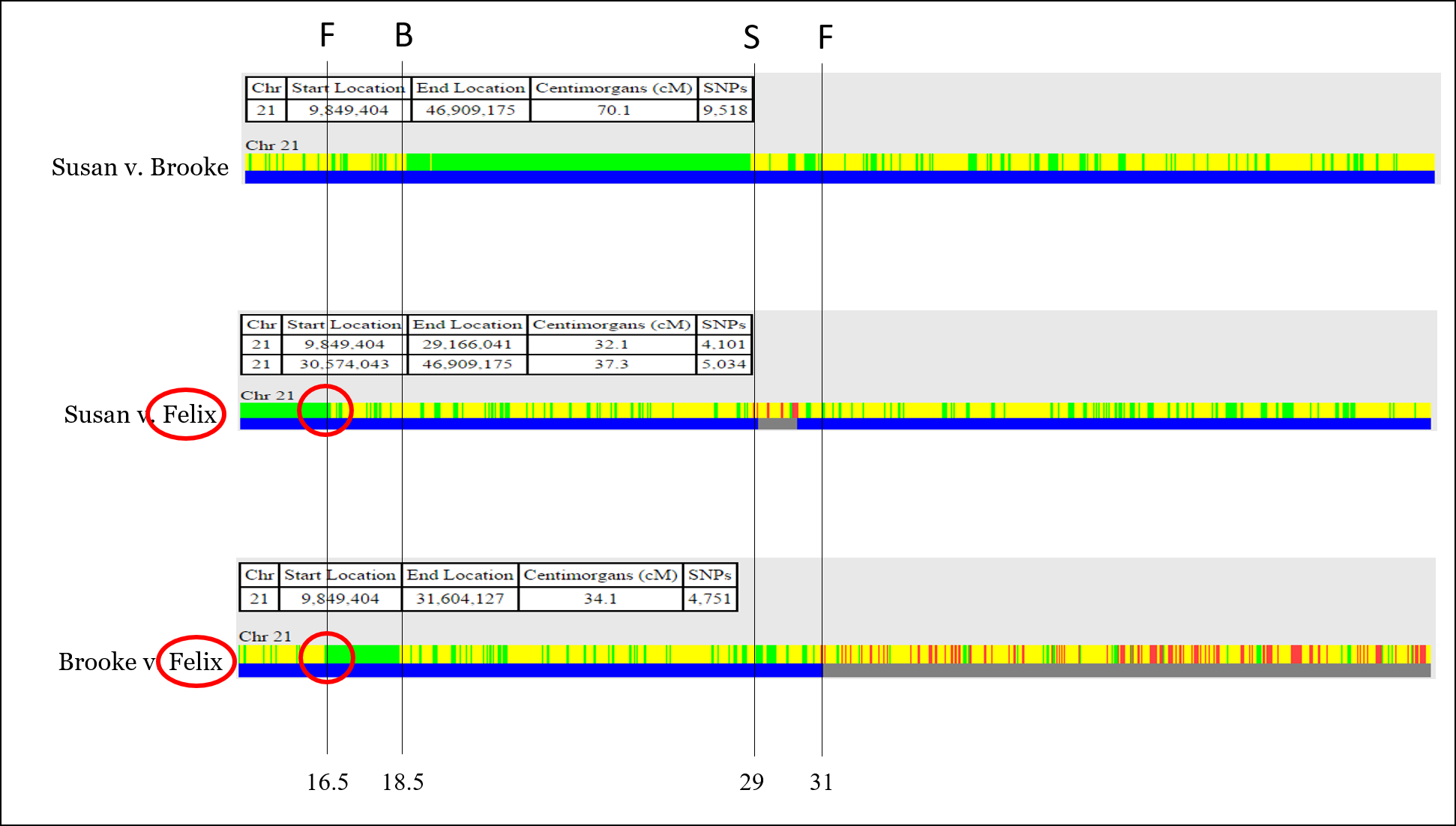
So now we have positions for each of the recombination points:
Now, let’s assign each recombination point to the person for which that recombination occurred. This is usually as straightforward as identifying which sibling has the recombination event twice. For example, Felix owns the first recombination point:
Now all recombination points are identified and labeled.
In the next post, we’ll start assigning segments of DNA based on the identified and labeled recombination points.


One of the aids I use in this process is to note the number of shared segments for each slice for each sibling.
From above
1 1 2 1 1 Felix
2 1 1 0 1 Brooke
1 2 1 1 0 Susan
I find it helpful to note when there is a 0-2 or 2-0 jump, i.e. when there are multiple recombination points near the same spot. This highlights the need to look more closely with Full Resolution to differentiate the more difficult to see recombination points.
Fascinating, Dave! The 0-2 or 2-0 transition is confusing the first time you see it, for sure. I’m convinced that this process could be automated, and your method seems like it could be part of that. Thank you for sharing!
Thanks, Blaine.
Another useful tool I was introduced to was David Pike’s utility http://www.math.mun.ca/~dapike/FF23utils/pair-comp.php which provides start and end points for the single and double match segments. The only downside relative to GedMatch is that it uses your raw data files which are generally Build 37 so the start and end points are off from GedMatch’s converted Build 36 points. They are generally close but it would be nice if GedMatch offered that tool so there would be no discrepancy. It could be part of their Tier 1 offering. I think this could bring things closer to an automated process.
I am thankful you and others have been working to popularize the 3 sibling grandparents phasing. It makes me feel like I am getting real value out of the DNA tests.
Could someone explain how Dave is counting the shared segments? It looks like for Felix, Dave is counting what is the double, single or no matches for Susan V. Brook. How does that count Felix’s segments?
Very useful, Blaine. Thank you.
Thank you, Bill! I’m glad you found it useful!
This is a powerful tool. After being introduced to it last summer by Blaine and CeCe at GRIP last summer, I undertook to verify that a match between my wife and her two siblings was actually through my wife’s maternal biological grandfather. With help from Kathy Johnston, I was able to convince myself that was the channel through whom the DNA was inherited.
A caveat: additional testing of cousins on the other side of this match subsequently has demonstrated that my wife’s maternal biological grandfather was not the presumed maternal grandfather.
Visual phasing was an important tool in helping penetrate this genomic puzzle. However, in our complex case it also took additional yDNA, atDNA and finally xDNA results beyond those originally involved in the visual phasing to have some confidence we have discovered the correct relationships.
Dave, I’m so glad you have used the methodology so successfully! I too view visual phasing as another piece of the overall puzzle.
Great post — I’m anxiously waiting for the next one so I can continue the process.
I have a question (probably a dumb one) about the process of determining the recombination points. You state that “Anywhere there is a change in the sharing status between two siblings, there must be a recombination event in at least ONE of the siblings (and sometimes both!).” So, for example, if it changes from yellow to green or vice versa, that must be a recombination event. Seems simple enough, but in your example, for Susan v. Brooke, the first recombination point is shown as the beginning of a large chunk of green. However, there are quite a few changes from yellow to green and back again in the area before that identified recombination point. My guess is that areas where the colors change back and forth in “small chunks” should be ignored; look for the beginning and end points of “big chunks” of color. If that is the case, my question is, how do you know for sure when to ignore the changes and when to count them?
No, there’s no such thing as dumb questions when it comes to visual phasing! It’s a great question.
Yes, you should ignore those very small segments. The yellow segments will always be identified by the blue bars. For the green segments (sharing on both chromosomes), focus on sustained green segments. There’s no exact guidance here, but generally you will be able to tell the difference between the long green segments and the smaller “spikes” of green sharing. The more sibling comparisons you see, the better your feel for the difference.
Thanks Blaine, that helps. My two siblings and I have all tested and are already uploaded to GEDMatch, so I’ll try this out and see what I get.
Will visual phasing work if one of the 3 siblings is a half sibling (we have the same father)?
The consensus is that it will, although of course you’ll only be phasing one side of the family (the paternal side) rather than both sides.
I recommend that you find a friend that has 3 siblings at GEDmatch you can work with first, to get a handle on the methodology, but after that I’d love to hear how you did with two full siblings and a half sibling!
Hmmm; I have half siblings on both maternal and paternal sides….what an interesting prospect. I do not have 3 full siblings. This site is great!!
I can’t wait to try this. I have a group of 8 siblings to practice with – thanks!!
8 siblings!? What fun!
Would analyzing four siblings be provide stronger or additional results?
Yes, although you’ll want to work with 3 siblings at a time. So you can do two sets of three, with two people overlapping. That 4th sibling can really help you figure out problem areas, especially.
My husband matches three siblings. Could I use this technique on the matching chromosomes to help me focus my research?
No, unfortunately. This technique is used to phase the chromosomes of three siblings themselves, not someone who matches three siblings.
Well, you could ask the manager of the three siblings to carry out this technique and report which grandparent matches your husband’s segment.
Maybe it is just late in the day, but as to line up the chromosome, are we stretching each chromosome so the length is the same? Does this counter act the image reduction note under each chromosome graph mentioned on Gedmatch?
Thanks.
Well, we’re only working with one chromosome at a time. So let’s say I’m working with chromosome 22, and I have three images: one for Brooke, one for Felix, and one for Susan. Sometimes, the images for one or two of the siblings isn’t exactly the same as the others, so I will stretch those out, but it is a very small, minor adjustment.
So, you’re not stretching chromosome 22 to match chromosome 1, for example, because they’re never compared to each other.
If you have 8 potential living siblings, is it worth it to test all 8?
That’s a tough one to answer. Having 4 or 5 will really enable you to visually phase your chromosome with a high degree of confidence. You’ll also have a lot of your grandparents’ DNA tested as well (unless you already have your parents tested?), which would be great if this process ever becomes a tool that enables us to recreate our grandparents’ genomes. Which I hope it might!
I’m starting with the X-chromosome comparing me with my two brothers and I’m getting confused. Just to clarify, if two brothers match completely on sections of the X-chromosome, does that mean that segment came from both of my maternal grandparents and on the sections where they don’t match at all (mostly red: base pairs with no match), is that where the segment comes from just one of my maternal grandparents and I have to figure out which one? I do have some cousins on that side to help figure out which is which. Thanks!
This is fun! I have four siblings – 1 female – 3 males. (and only one of their parents).
You suggestion to start with the X makes sense. Question: would you recommend grouping the three males together as one group and including in the girl in the second overlapping group? Or do you think having the female in both groups is better? I also then have the maternal next generation up with two females and two males. My gut says it may help to figure it out with having the two males in one group and one of the females and then overlapping with the second female and the same two males. Any thoughts? This is cool because we now can help potentially to ID maternal GGP’s for the first round! Thanks Blaine!
Is there a way to do this if there are only 2 siblings? Will we get at least an approximation? Would using a cousin as the third help in any way?
Wow! Thanks for this great article. It is particularly helpful for me because I have already done DNA tests on my mom and two of her siblings and all three of them are on GEDmatch.com. Now it’s just up to me to do the process. I’ve really been anxious to get one of their first cousins tested because her father was the identical twin of my mom’s father. Genetically a half-sister.
I do have a question. My dad’s parents were 1C1R with lots of endogamy in their family tree. He still has 4 remaining siblings. Would this process be as effective with that group as well?
Hello Blaine
I recently had the third sibling of my mother tested and was excited to be able to utilise your suggestions about visual phasing. As you suggested I started with X and have several great clues emerging as a result.
I am now working on chromosome 17 which was a bit more tricky but I think my output is reasonable and seems to be working out as expected except for one small segment area (I have close paternal and maternal cousins for them that has helped test the theory). According to my 4 grandparent segments, in this particular area all 3 siblings share the segments from the same grandparent, one for both sides.
I have a few matches that only match my Mum. These matches also match me, my son and my phased maternal kit, so they look IBD.
So my question is, what is more likely? Are these matches more likely to be IBS, or have I got the phasing wrong?
Veronica
I have been working on doing the visual phasing for both my father’s and mother’s side of the family for several months now. I am fortunate that Randy is the leader of the DNA SIG of my local society, so I’ve been using his methodology on the project. My goal is to derived the half profile of each of my great grandparents (i.e. the part they passed on to my grandparents) using this method and then use cross comparisons of other known cousins I have tested to start completing the other half.
For my paternal project I have tested my father and each of his 3 living brothers and both children of a brother who died. My mother’s family is even larger. I now have the DNA from my mother and 4 of her siblings and from two first cousins, each through one of the siblings that hasn’t been tested (giving me DNA segments from 7 of the 9 surviving lines). Additional testing on that side is underway to plug a few holes in my grandparents’ profiles where the tested siblings each got the same maternal or paternal chromosome, but the grandchildren from the untested siblings appear to have part of it. Testing the siblings may fill in the gap.
I’ve learned from my months of working on this personal project and have some starting off advice for those considering undertaking this method as a project:
1) It helps in the alignment of the crossover points in the GEDmatch pairwise comparisons if all the testing is done with one company. Because of the way GEDmatch draws the one-to-one comparisons, if Person A and Person B have radically different locations on a specific chromosome that were tested by different companies, it can throw off the alignment, sometimes quick dramatically, of the crossover points. Remember that FTDNA’s Family Finder test and the Ancestry DNA test version 2 now only share about 420,000 common locations instead of the approximately 660,000 of the version 1 test. And 23 and Me’s test, depending on version, also varies greatly in the locations tested compared to Family Finder and Ancestry DNA. The new full resolution option helps quite a bit, but it is still a work in progress. This also comes into play if you are doing reconstructive DNA profiles based on the mapping. You want as many locations in the DNA profiles where the siblings all had that location tested at their testing company.
2) While you generally need three siblings to start, it is also beneficial to have a few people the next generation down too (whether they are from a person already tested or from a sibling no longer available) in order to act as “tiebreakers” in several instances, most notably when you have one set of siblings who are FID to each other in one region and all the remaining siblings are FID to each other, but non-identical to the first set. Figuring out whether it was a paternal or maternal chromosome that flipped at the crossover point can be problematic. If you are taking the project to the level of actually developing a profile of the parents or grandparents of the siblings, you will need these children/nieces/nephews of the sibling’s generation to be able to harvest the SNPs in these FID/NID regions. Otherwise you will only be able to harvest the SNPs that are homozygous, which when you upload the profile to GEDmatch, will cause the two unrelated grandparents to appear related in that region.
3) Watch out for multiple crossover points bunched together. I’ve had the instance where three of the siblings each had a crossover point within 8 tested base pairs. Having as many siblings tested as are available will help in these instances. Having multiple close crossovers will also result in GEDmatch not always catching a HID or FID region and labeling it in blue in the lower part of the one-to-one comparison. Be on the lookout for these. I’ve found lowering the thresholds to the 400 to 450 SNP range as opposed to the 700 SNP default helps, but can occasionally produce a false positive too. Since you are comparing siblings, lowering the thresholds in this manner is not as dangerous as it would be for comparing more distantly related persons.
Thanks for the advice about bunched multiple crossover points. I’ve been wavering as to whether these are real, or just the “fuzziness” mentioned in the description of the method. So far, I’ve just worked on four very short chromosomes, and I’ve already found several instances where GEDmatch does not seem to be catching short HID or FID regions. I’ve been ignoring these, but now I think I need to go back and take them into account. (I’m using four siblings — myself and three brothers — and using my mother’s half uncle, my father’s first cousin, and other matches with known MRCAs to help identify the grandparents.) However, I’m afraid my head might explode!
Will this work if one of the siblings is a half?
Hi Blaine,
Would this method help us determine which of two brothers (both deceased) was our paternal biological grandfather? We have tested some great grandchildren of one of these brothers but the other brother in question has no descendants to test. Three of my siblings have tested and I have several more that are willing to be tested. Our father is deceased.
Correcton to my previous email: Sorry Blaine….In my last email, I incorrectly identified the descendants of one of the brothers that could be our paternal grandfather as “Great grandchildren” but they are actually GRANDCHILDREN of that brother. Also, the two brothers in question had another brother (not in question) and we have tested both his daughter and grand-daughter. Would visual phasing help us distinguish which of these brothers was our paternal grandfather?
Thanks so much!!!
Dear Blaine!
Thanks for your extensive explanation. Unfortunately, I have just one sibling (brother). My father also has one brother, who has 2 children. My mother has a half-sister, who has 2 children. I also have my maternal grandmother still alive.
Is any combination of these relatives suitable for visual phasing?
Thanks,
Mantvydas
Three siblings and myself have had our DNA tested. Also, my mother’s brother and my cousin on my mother’s side. We are searching for the unknownn identity of our maternal grt, grandparents! Would phasing help discover who they were?
Where can I find Part III?
This methodology is fascinating. I wonder if it could help me with my puzzle.
When I tested my ethnicity estimate said I was 10% western European Jewish. Since I have my paperwork on my ancestors done back to the 2g grandparents with no one of identifiable Jewish ancestry in the picture, I passed this off as a curiosity of the ethnicity estimating algorithm. Then my cousin tested (the son of my mother’s brother) and he got the same result, 9% western European Jewish. Then I got my aunt (the sister of my mother and his father) to test and she came back with 20% Jewish. So it seems the Jewish heritage is real and it is coming to me down my mother’s side. From these numbers it appears to me that a likely scenario is that one of my great grandparents on my mother’s side was Jewish.
This means that one of my great grandmothers probably conceived my grandfather or my grandmother with someone other than her husband. Since I know already that each of these women had other children with men not their husbands, it is not hard to imagine that this might be so.
My question is, if I can successfully do this chromosome mapping exercise so as to see from which grandparent I got my chromosomes, will I then be able to see from which grandparent’s chromosomes the “Jewish SNPs” derive?
Thanks for your thoughts.
Gedmatch has a checkbox at the bottom of the 1:1 comparison to Show only Full-Match (FIR) segments.
Clicking on this will give the start and stop of the green segments.