The Shared cM Project is a collaborative data collection and analysis project created to understand the ranges of shared centiMorgans associated with various known relationships. As of August 2017, total shared cM data for more than 25,000 known relationships has been provided. To add your data, the Submission Portal is HERE. I am always collecting data, and perhaps the next update with have 50,000 or 100,000 relationships!
This August 2017 update is the second update to the original data, released in May 2015, and includes many thousands of new submissions.
There is MUCH more about the project, including histograms and company breakdowns in the PDF download.
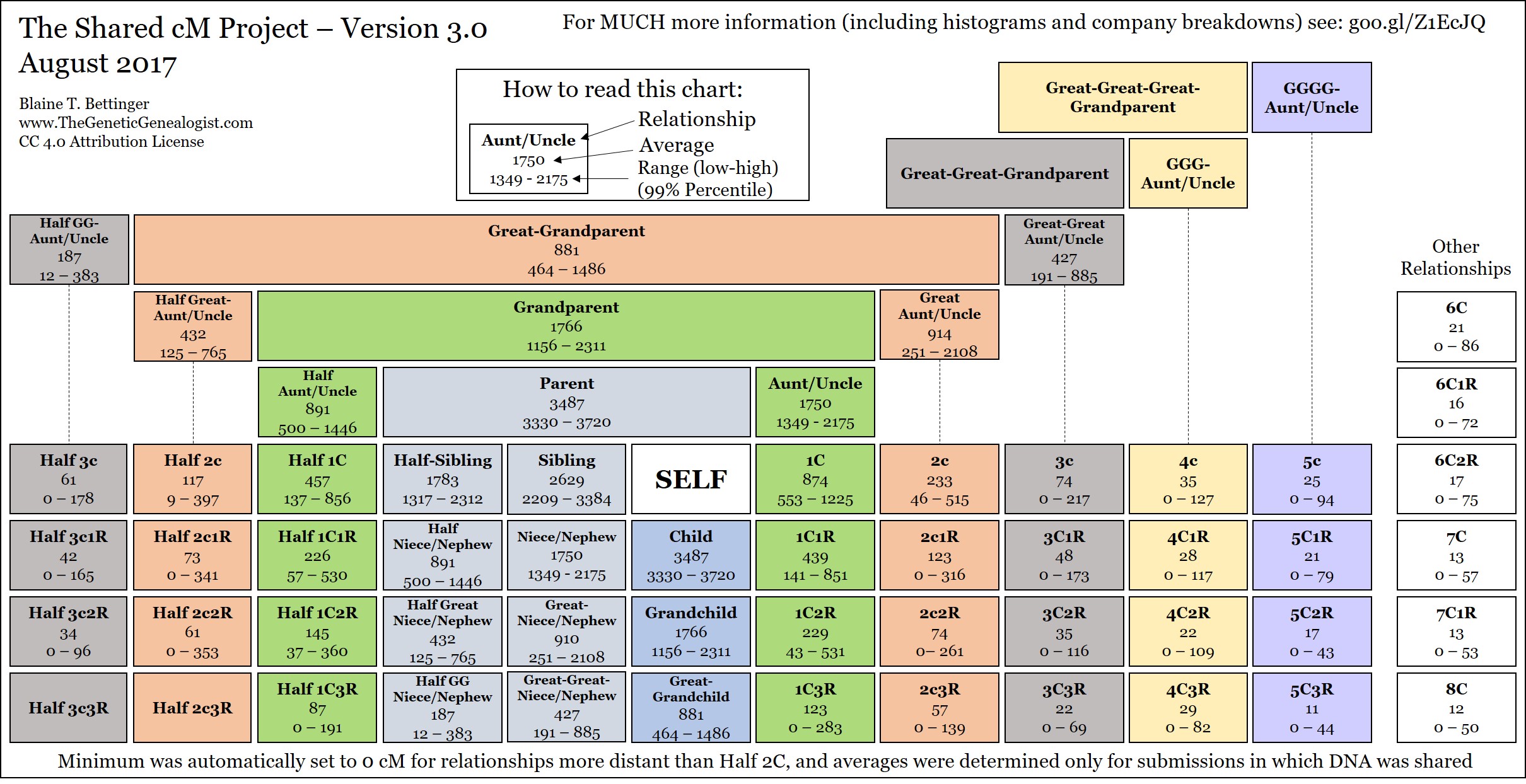
Figure 1. The Relationship Chart
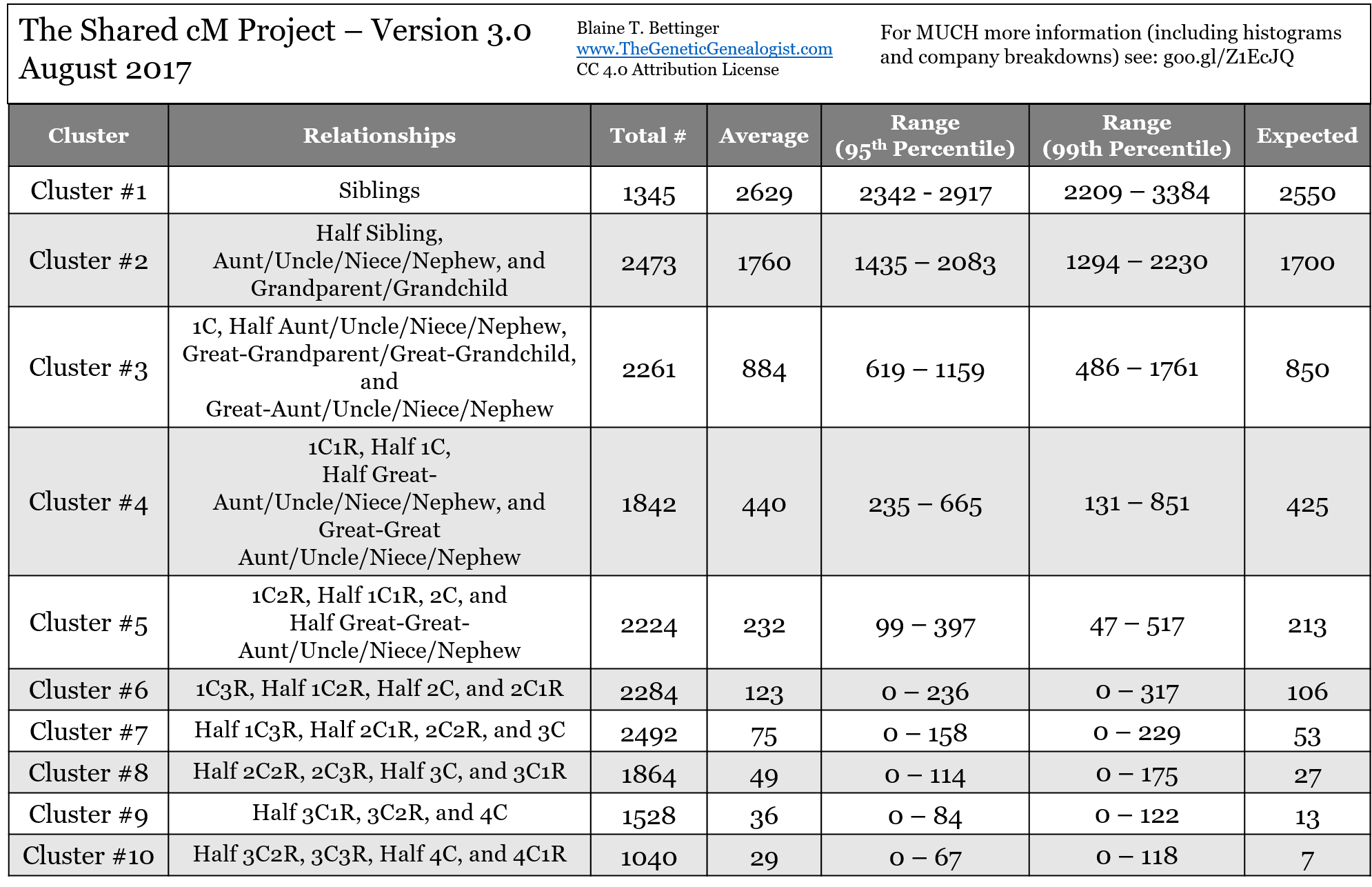
Table 1. The Cluster Chart
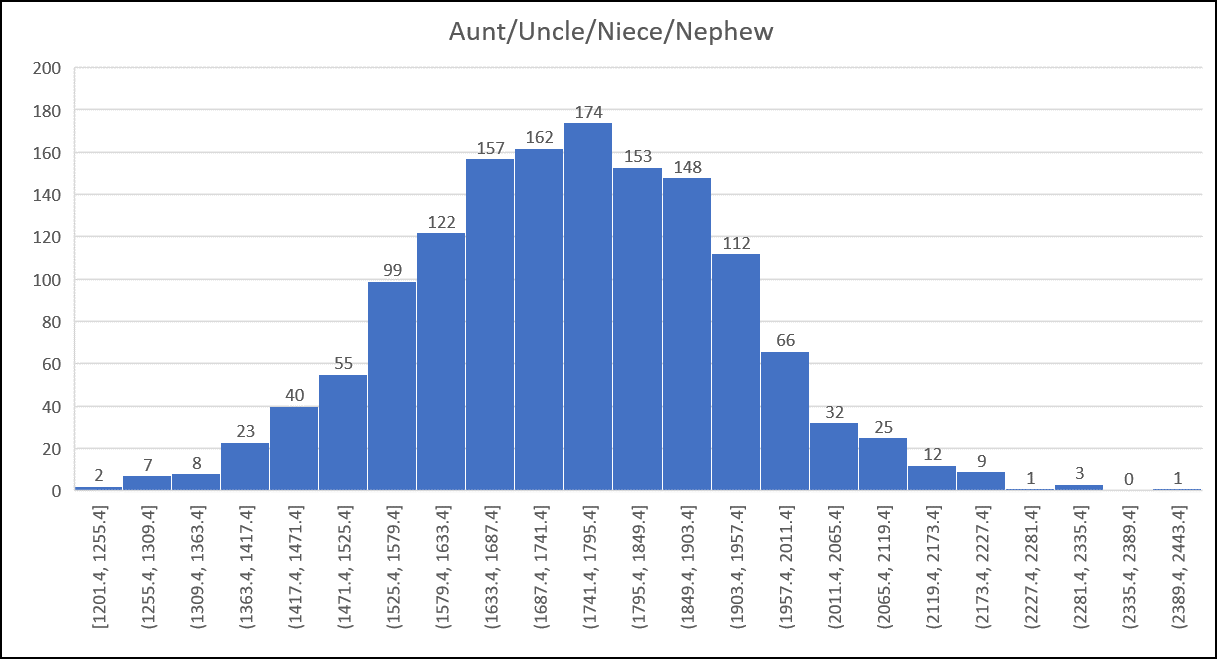
Sample Histogram from the Shared cM Project (all histograms available in the PDF download):
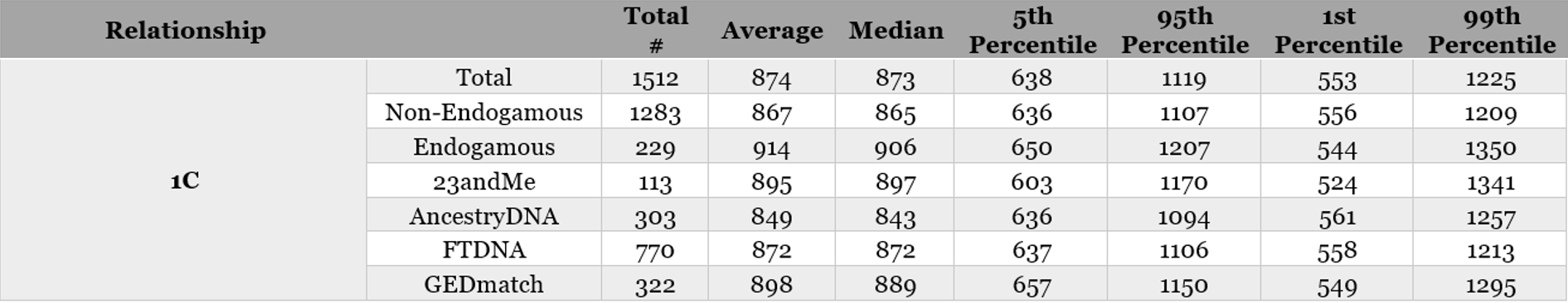
Sample of Table 3. Analysis of endogamy and company breakdown for 1C (all company breakdowns available in the PDF download):
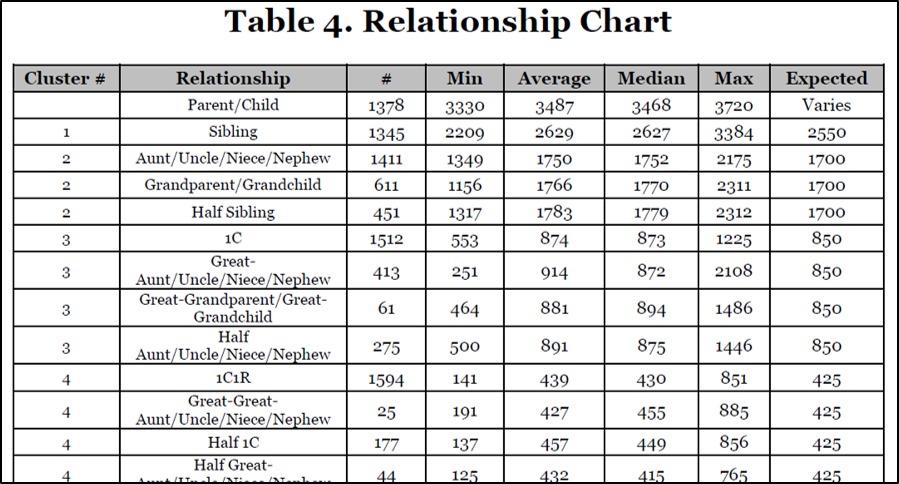
Sample from Table 4:
.
Wow! What a tremendous amount of work. Thank you, Blaine! I used your charts all the time.
I have a lot of known cousins now whom I want to enter as data points into your project. But…I do not remember exactly whom I have already entered (very few I think). Will you be able to weed out duplicate entries (maybe based on my email address)? I will keep track now of whom I enter…
An invaluable resource, THANK YOU!
I haven’t read the pdf yet, but is there not enough significant difference between testing company results to warrant additional charts?
Ignore my question (not my thanks though). Some day I’ll learn to read a bit more before I post, lol.
I frequently refer your chart to users of GEDmatch. This update will be a big help. It is an invaluable aid to the genealogical community and an instant classic tool .
Thank you so much for the time and effort you spend generating these references for the community.
I have put your chart on my phone! Thank you. I have one question about the average for the 4c1r, is it supposed to be 29? That makes it higher than the 4c1r. Thank you!
We reported a 480 from FT for cluster #3 – which is literally off your chart! (We have no reason to suspect anything non-parental.) Gedmatch gives the same pair a sightly higher 541cM.
aunt-nephew M-J 1,552 longest block 140 (gedmatch:1685.8 133.9) cluster 2
father-son J-D 3,383 longest block 267 (gedmatch:3586.5 281.5) cluster 1
greataunt-greatnephew M-D 480 longest block 72 (gedmatch:541.2 71.6) cluster 3
As you noted in your PDF, different testing companies calculate the total shared cM differently, depending on the minimum size segments that they include in that total. What minimum size did YOU use when creating this chart?
Thank you very much for this extremely valuable resource.
Are you on GedMatch.com? Kathleen HURLEY Doan
Love your book The Family Tree Guide to DNA testing an Genetic Genealogy. I have entered several people I’m not sure if I entered all of them. When you get 50% of Father and 50% of Mother’s dna this is just an average right. How much can this vary? In the book between grandparent and 2 grandson’s the difference 22-28 for 1 and 17.7-32.3 for the other. Judy
I don’t see any replies on here from the author but hopefully they are reading. I didn’t see any spot on the interactive chart for identical twin result. Identical twin is confusing as it appears to be the same cM’s as parent/child. You would think it would be much more. An identical twin article would be an interesting read for me and maybe others
I too am desperate for more information on children of identical twins…
Dear Blaine, thanks again for the great work. I am presently investigating the match to me, on AncestryDNA, of a half nephew (son of a half sister). Am I correct in seeing minor differences in the numbers on the Relationship Chart vs. the Cluster Chart? The Relationship Chart states that, for the relationship of half nephew, the average amount of shared cM was 891 and the range was from 500-1446 cM. relationship chart was 891 and the range was from 500-1446 cM. The cluster chart says for cluster 3 relationships the average is 884 and the range 619-1159. Am I missing something? Thank you
Blaine,
Great work with the shared cm segments because it is essential. We have a very valuable tool (autosomal dna testing), but the statistical nature of the results make it nearly unusable except for the closest matches. An accurate, well prepared genealogy is absolutely necessary to make sense of the results for more distant matches. The hope is that your work will refine the statistics for more distant matches. However, there are several assumptions that must be stated.
The first assumption is that the genealogies used to determine our degree of relationship are accurate and well prepared. I will not use family trees from some genealogical websites because I am not always able to confirm the provided information. This problem becomes quite important when attempting to differentiate at some levels of relationship (for example 8th cousins and 8th cousins once removed).
The second assumption is related to the first. For the statistics to be valid the two related persons must be related in only one way. This becomes less likely at more distant generations but is harder to prove. This would require knowing all 512 persons in our 10th generation (8th cousins) to provide valid information for your tables.
The third assumption is that we all understand endogamy the same way. Incest and marriage not permitted by a church are obvious, but what of 2nd or 3rd cousin marriages? Second and third cousin marriages have a measurable and not insignificant impact on the collected evidence. Similarly, fourth and fifth cousin marriages have a measurable, but smaller, impact on the statics. Unfortunately, the last two examples are harder to prove without an extensive and accurate family tree. This is extremely important at the most distant relationships where the uncertainty, as currently reported, far exceeds the measured or predicted value.
The assumptions mentioned must be evaluated for every set of data submitted.
I would be very interested in getting the percent for each cluster in small ranges of shared cM. Like fore the range 440cM-460cM: 2% cluster #3, 87% cluster #4 and 11% cluster #5. This would then easily be compared to this table https://i2.wp.com/thednageek.com/wp-content/uploads/2017/01/AncestryDNAs-Figure-5.2-Table-of-probabilities-2-1.png from the simulation done by AncestryDNA.
http://thednageek.com/the-limits-of-predicting-relationships-using-dna/
https://www.ancestry.com/dna/resource/whitePaper/AncestryDNA-Matching-White-Paper
Hi,
I recently received the results of a great-grandmother of mine on ancestry. We share 456 centimorgans on ancestry and 488 on gedmatch. We also have no shared matches on ancestry. This is substantially lower than the centimorgans I share with another great-grandmother of mine with whom I share 989 centimorgans on ancestry and 1157 on gedmatch. Why is this?
It is essential for science to state the assumptions and parameters used.
So why is it not stated in the article, in the PDF, or in the portal whether X-DNA is considered in these numbers?
At gedmatch.com, the major crossroads of amateur DNA study, there is a cM given for autosomal DNA, or a cM for X-DNA. You can click “A” to get a one-to-one autosomal comparison, or you can click “X” to get a one-to-one X-DNA comparison. There is no letter you can click to get a full one-to-one DNA comparison. You have to do the addition yourself. Instead of stating this parameter, the PDF, this article, and the portal studiously avoid the use of both “autosomal” and of “x-dna” … except on page 4 of the PDF where a click to a document https://isogg.org/wiki/Autosomal_DNA_statistics with “autosomal” in the title produces 34 instances of the word “autosomal”. Right near the top, we learn that “autosomal” excludes X-DNA with this statement: “Autosomal DNA is inherited equally from both parents.” So while the general theme is that the study deals with 1-23, the detail points to 1-22.
The portal has no method to exclude duplicate reporting of results, no checking, and is ambiguous on what it is requesting. So kudos for the effort, but I believe that the methodology is irredeemably flawed. You need to start over.
I posted the above a few days ago, but it disappeared without explanation. 11 Jan 2018.
I am using your portal to enter my data for your research. I have used 23andMe.
How do I locate the longest block of DNA in cM for my entries? Is it available for that
site? Thanks.
Curious, the averages you have for the more distant cousins presumably exclude zeroes, right? I mean, unless you make a probabilistic assumption about how many 6th cousins tested (for example) I don’t know how you’d know how many zeroes there were.
Does the total amount of centiMorgans shown in the Shared cM Project at each relationship include the both the 22 autosomes and the X chromosome, or just the amount for the 22 autosomes?
It is not clear from the website or the instructions for submitting. If it is stated can you please diret me to thre relevant text.
Many thanks.
Hi Alasdair!
Report whatever the company reports to you, don’t anything else. Some companies report X, some don’t, but it’s already either included in the total or it isn’t. So no need to worry about any extra steps. Thanks for submitting!
First off – thanks. You have created a reference oint for everyone wondering how their “DNA matches” might plausibly be related to them.
I have a couple of questions on methodology – others have been raised before in this feed (one a rather misdirected rant), the second on how to treat no-matches when giving average results I think is more problematic.
My first question is how do you compensate for the vastly different matching results from different companies? I see you request the test provider information on your submission sheet – but do you try to allow for the differences between organisations and companies providing conservative matching values (GEDmatch, 23andMe, Ancestry) against those that give more liberal estimates (FTDNA, My Heritage)?
Second question relates to submission bias. It is not more tempting to add results that are outliers than it is to dilligently enter every known relationship? Just a thought.
Whether you think my points are valid or not, I salute you on your project.
1. The PDF contains a breakdown for each company (other than MyHeritage, which is too new to have enough submissions in the project). The variations are not as great as one would think, and of course they’re all just a subset of the full variation for each relationship.
2. Submission bias is definitely an issue, as is data entry and other factors discussed in the methodology section of the PDF. But this is why scientists and statisticians love big data. The more data we receive for the project, the lower the probability that these are swinging the data significantly. For example, there are >1,500 submissions for 1C. The likelihood that submission bias is significantly biasing the data is very low.
Hi Blaine,
Just wondering what settings you suggest for GEDmatch for one to one compare? Do we leave the settings as default or increase the threshold for segment size to 10cM?
Thank you in advance.
I submitted data for a 2nd cousin, but i have my matches separated into 7+cMs and 7 category. Can I correct this, or is this the way you want the data?
Thanks.
Sorry, only part of what I wrote came through. The data I submitted for this 2nd cousin only included the 7+cM segments, but there are two smaller segments, so the total cM and the number of segments are greater than what I submitted. Is this what you want, or do you want all of the segments?
Thanks again.
Question about twins.
I notice that the range for siblings is 2209 – 3384.
Elsewhere I’ve seen identical twins are indicated as sharing 3400.
If two twins come back as 3384, does this mean they aren’t identical, but instead fraternal sharing the absolute maximum number of centimorgans that is possible for siblings and still not be identical?
Hello,
This is great work! I made a simulation that can predict the amount of shared DNA between people. The only problem is that my predictions are an average based on the assumption that there’s no difference between the ways that men and women recombine chromosomes. Since DNA from fathers is recombined much less, the variance is much greater in shared DNA through paternal lines. I’m wondering if you have statistics on shared DNA between grandparents and their grandchildren in which the sex of both the grandparent and the parent are known–really just the standard deviation or variance would be great.
Thank you,
Brit
Great job, Blane! Thank you. I used your charts many times. Also there are a lot of information now which I want to recommend on https://pro-papers.com/gb/genetics-writing-service as data points into your project. I’m also wondering about statistics on shared DNA between grandparents and their grandchildren. If you have, it will be nice to see.
Please HELP! I’ve been looking for my bio dad for many years. I was linked with a 1st cousin which is not related to my mother’s side. However, she only had 3 uncles all died. All had children and were tested. 2 of them came up 1st cousins with cMs ranging from 839 to 737 across at least 36 segments. One came up with 1170 cMs across 51 segments. She is the only option as my half-sister. Everyone tested was female including me if that matters. The question can she be my half-sister with an 1170 cMs? Tests were done via Ancestry
There’s only an 8.71% chance that she’s your half-sibling and a 91.29% that she’s your first cousin when you put the Shared cM Project 3.0 tool v4 (https://dnapainter.com/tools/sharedcmv4)
Have you tried uploading all the DNA tests to GEDmatch and comparing them in depth?
Hi: I gave birth to two children. Both of them have done the ancestry DNA test as have 1. However, I noticed something strange my 1st child (female) shows 3,275 shared cMs, but my 2nd child (male) shows more: 3,362 cMs. They are full siblings. Why is there such a difference in the cMs? Also, if a person supposedly has 6800 cMs, why do both of them show less than 3400? According to the charts, they’re not my offspring but possibly my siblings…and I KNOW that can’t be right as I clearly remember going into labor and giving both to both kids lol. At any rate, I don’t like the idea that my results don’t look like normal results. Is it possible that Ancestry dot com made a mistake on the number? mind, they identify us all as parent/child but yet all the other cMs charts online show otherwise. Starting to really distress me.
Maria, I’m not an expert or a professional, but I think it’s just a quirk of AncestryDNA’s algorithms and how they are interpreting your DNA data, and I wouldn’t worry about it.
I haven’t done the DNA test, but my son and parents have. I’ve downloaded their data from Ancestry and uploaded it to GEDmatch and FTDNA, and they all show different cM figures.
For my son and my dad: AncestryDNA says 1993 cM; GEDmatch says 2052.2 cM; FTDNA says 1919 cM.
For my son and my mom: AncestryDNA says 1506 cM; GEDmatch says 1557.7 cM; FTDNA says 1479 cM.
Theoretically, if you total the cM my son matches w/my mom and dad, we would know the cM I share with my son. If so, Ancestry DNA puts us at 3499, GEDmatch puts us at 3609.9, and FTDNA puts us at 3398.
Clearly, the amount of DNA my son shares with my parents in total has and always will be 50%, regardless of how the companies interpret their DNA data. Ultimately, I think that the differences in the figures are due to the fact that a “centiMorgan” is, essentially, a theoretical thing being measured in a theoretical way, so the measurements are not exact. It just seems to us like they SHOULD be exact because we are used to measuring physical things in a physical way, like with a ruler or tape measure.
Genetic shuffling during the creation of the sex cells (sperm and egg) is a random process, with natural variation. The DNA from both sets of chromosomes lines up and breaks in the DNA allow a re-shuffling of the original DNA that you inherited from your parents! So each sperm and egg have a slightly different result from the re-shuffling process. These variations are not unusual for full siblings. You have identified two data points from a wide distribution of the possible outcomes from such a process. Rest assured.
Just 2 suggestions:
1. in the self square, add the number of cM
2. list that cM are centiMorgans
I have a first cousin match on GED Match sharing 883.4 CM, largest segment is 73.5 cM 28 shared segments Autosomal one to one comparison . Our GED Match X DNA One to One comparison shows match of 177.0 cM largest segment being 95.5 cM 93.156 percent . We are both females same generation is it possible our fathers are brothers. She knows how her father is , I do not .
Is Figure 1. The Relationship Chart complete? My Great-Aunt son would be my 1st Cousin once-Removed. I do not see the averages for that relationship.
I’ve been running around the net trying to find an answer to my question.
I have found a half brother 1,299 cm longest block 189
half niece 1,256 131
23andme and family finder have given me the relationship range HOWEVER, according to your charts I’m a bit short on Centimorgans to have either of these relationships.
Jeannie – you may find this helpful: https://dnapainter.com/tools/sharedcmv4/1299
Greetings Blaine. Thanks very much for this resource. I have a question about tools for using the results of multiple DNA tests to solve genetic relationship questions from a couple of generations back where uncertainty is greater. Presumably from a statistical perspective, cM data from multiple descendants might tighten confidence when trying to make inferences about, for example, which of several individuals is a likely biological ancestor? I am just curious to know if you are aware of anyone developing such a tool?
This is a great resource, thank you. Feature request: please show standard deviation in your tables. If I understand the methodology min/max are determined by one or a few outliers so somewhat arbitrary, whereas SD by nature is a statistic of the entire data set and so more representative. It is also useful because in a single statistic it expresses the spread of the data. Thank you!
Planned for next version hopefully. This isn’t SD, but I hope you are using the histograms! https://thegeneticgenealogist.com/wp-content/uploads/2017/08/Shared_cM_Project_2017.pdf
I am very skeptical about Blaire’s statements, since the probabilitys are unclear.
There is a theoretical chance of 2.3 for 3rd Cousins; 0.10 for 2nd Cousins once removed and for 2nd Cousins the probalitys of no dectable DNA Relationship increase to 1 to 9 Mio – that’s mathematically impossible.
Great post.
Wow, Blane, I am really impressed! Thank you so much for the information.
I was actually looking for statistics on DNA for grandparents and grandchildren. But I guess I could just use the available one, will it be possible to analyze the data to write my essay ( I am following the pints at https://paperhacker.com/how-to-write-a-good-essay ) Once again, thank you so much!
I’ve read the article with wonderful satisfaction and also could know something new I will use
for your own additional requirements.
Thanks for sharing this info, I really enjoyed your storytelling fashion.
Thank you for creating high-quality content.
There’s so much info and much more advertising online that
it is really tough to locate worthy and relevant info.
Now, nobody visits libraries where you are able to find primary sources.
In the digital world, you perform distinct
Many times I have bought essays on https://elitewritings.com/. I think it gives you the opportunity to save your time. Many of my friends also buy essays.
Hi Blaine, I’m a bit confused between the relationship chart and the cluster chart. My 1stC1R is coming up at 794cm, quite high. On the cluster chart he can only be 1C but in the relationship chart he can be in either range, am I reading this correctly, thanks
I would like to see a list of references in the scientific literature that explains the basics and basis of the use of cM’s.
In Figure 1. Relationship Chart. I think the values for 3C3R and 4C3R are reversed?
Blaine,
Could you please explain the gaps? i.e. If I am looking for a match to 650cM who is suppose to be my Grandfathers niece I don’t see where that fits into the chart. Looks like there are gaps where I would be looking for her.