OK, that could be one of the worst blog titles I’ve written, but it’s intentional. When people share this post, I want the title to clearly convey the lesson.
Small Segments are Poison
We know that many small segments are false, and thus that many distant matches are false positives. I have written about small segments and distant matches many times. For a few background articles, see the following:
- “Small Matching Segments – Friend or Foe?“
- “The Danger of Distant Matches“
- “The Effect of Phasing on Reducing False Distant Matches (Or, Phasing a Parent Using GEDmatch)“
The (most current as of September 2017) definitive article on the nature of false versus true small segments is “Reducing Pervasive False-Positive Identical-by-Descent Segments Detected by Large-Scale Pedigree Analysis.” The paper is available online for free (http://mbe.oxfordjournals.org/content/31/8/2212). In the paper, the researchers found that more than 67% of all reported segments shorter than 4 cM are false-positive segments. At least 60% of 4cM segments were false-positive, and at least 33% of 5 cM segments were false-positive. The number of false-positives decreased fairly rapidly above 5 cM. See my analysis of this paper here.
Additionally, I’ve found that 32% of my matches are not shared by either parent. See more here. And see a concise summary of similar analysis and blog posts by Debbie Kennett at “Comparing parent and child matches at AncestryDNA.”
Accordingly, I use “poison M&Ms” as an analogy (copied from my own comment at WikiTree):
I consider small segments to be “poison,” in that too many of them are false matches and we can’t tell the difference between the false segments and the real segments. I use “Poison M&Ms” as an example. If I handed someone a bowl of M&Ms and told them that 30% are poisoned and there’s no visual difference (similar to 30% of small segments of 5 cM or smaller), no one would eat the M&Ms. Similarly, we can’t use small segments because they poison our genealogical conclusions.
If the best science we have suggests that a significant percentage of small segments are false, and these small segments are not labeled “FALSE” or “TRUE,” is there any way we can use these segments? Or does the fact that so many of these segments are false poison all small segments?
Can We Use Small Segments?
Unfortunately, there is a pervasive belief in the genealogical field that there are ways to use small segments. The following are the two most common hypotheses I see:
- Hypothesis #1 – Sharing one or more large segments with a match means that the small segments shared with that match are real segments; AND/OR
- Hypothesis #2 – Knowing the genealogical connection with a match means that the small segments shared with that match are real segments.
However, there is (currently as of September 2017) no evidence that these hypotheses are correct, and some evidence that they are not correct.
Using Small Segments – A Case Study
So let’s test these hypotheses with a case study. To be clear a case study is only anecdote, but we can draw some hypotheses from the case study, and perhaps it will lead to a larger study.
In this example, I am using a cluster of individuals with known genealogical relationships (see the chart below) that share both large segments and small segments. Accordingly, I am testing both Hypothesis #1 and #2.
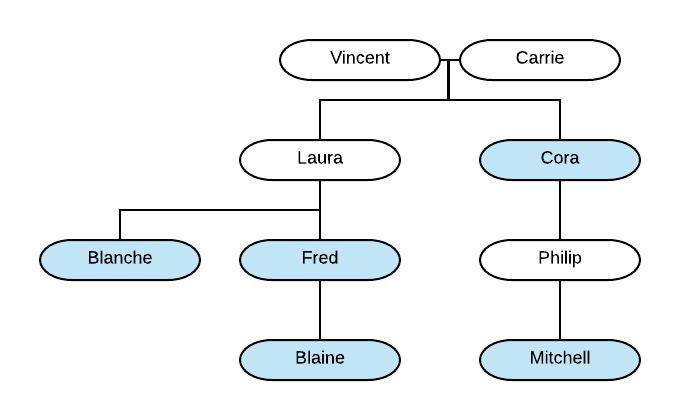
Blaine and Mitchell are Second Cousins. They’ve both tested at AncestryDNA and have transferred to GEDmatch. Doing a default One-to-One comparison at GEDmatch (with SNP threshold of 500 and cM threshold of 7 cM), they share the following segments:
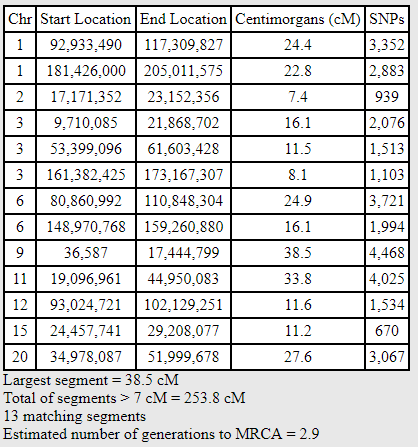
Total sharing of 253.8 cM is perfectly in the range for Second Cousins (see “August 2017 Update to the Shared cM Project“). The smallest segment is 7.4 cM.
If I lower the thresholds for the One-to-One Comparison to 250 SNPs and 3 cM, we share the following segments:
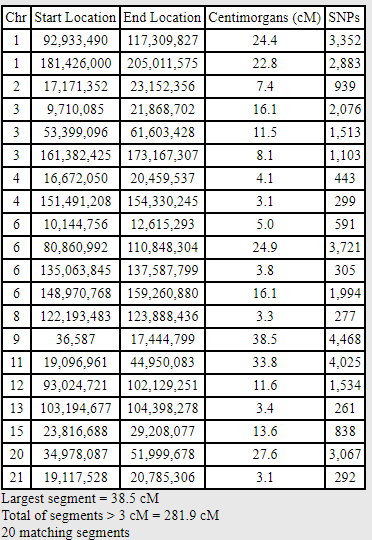
Because I lowered the thresholds, there are now 7 segments of 5 cM or smaller.
Are these valid segments? After all, there’s a known genealogical relationship, and I share many large segments with this match.
I have the advantage of being able to phase my results, as I’ve tested both my parents. When I compare my phased paternal kit to Mitchell using the same thresholds (SNP = 250 and cM = 3), I see the following segments:
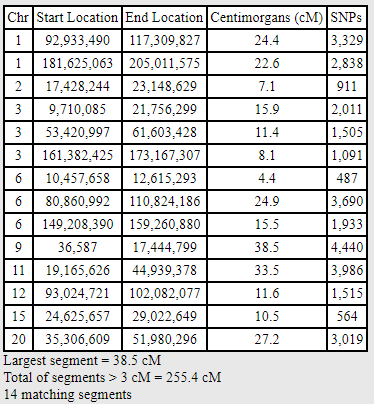
With one exception, each of the small segments has disappeared. Phasing, therefore, eliminated almost all the small segments despite the known relationship and the existence of large matches.
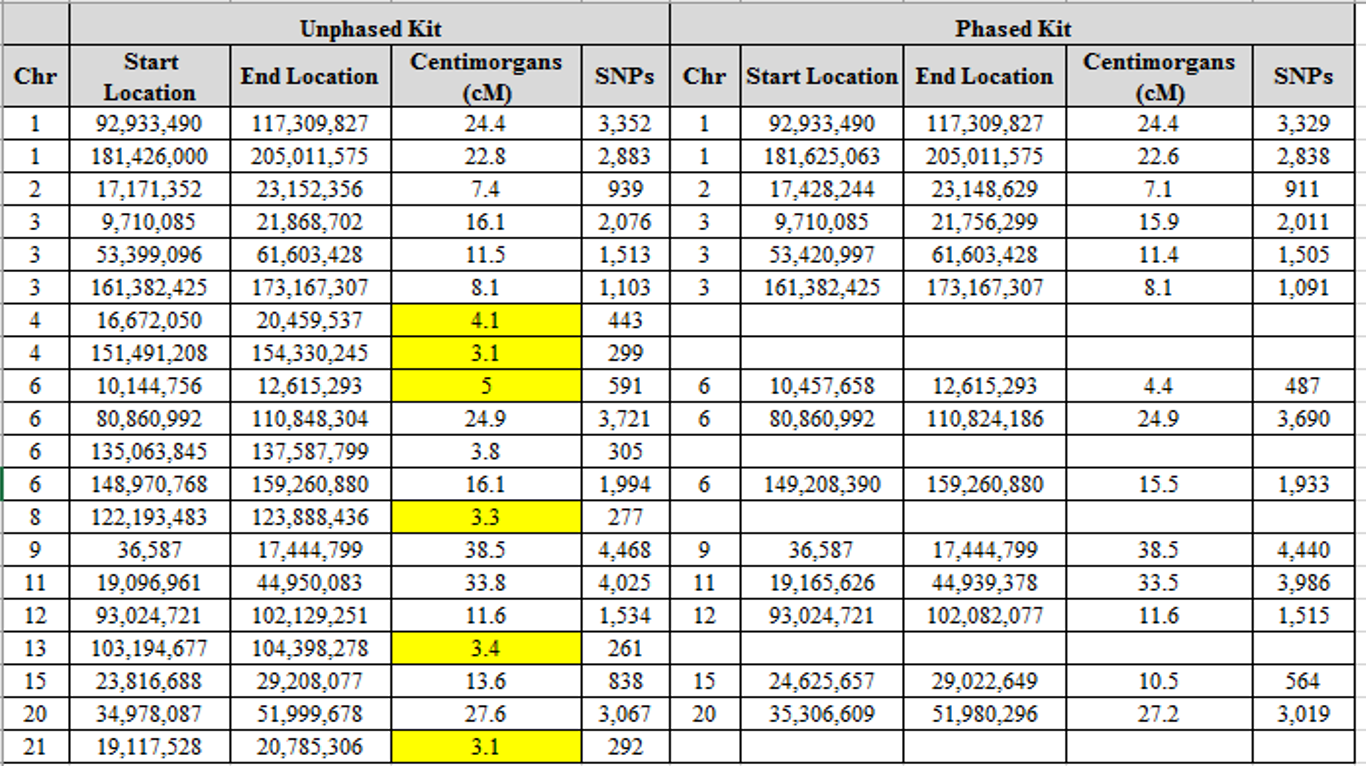
The 4.4 cM segment that remains may or may not be a valid segment (see below), but the point is that many or most of the small segments are eliminated by phasing. Unfortunately, the vast majority of people are not using phased kits when analyzing segments, usually because they cannot do so.
Note that I also compared my maternal phased kit to Mitchell using the same thresholds, and there were no segment shared; so these are truly false segments and not maternal segments.
This analysis also overlooks the fact that small segments are likely to be very old, and therefore could have been inherited from any of a number of possible lines, which may or may not be the line we’ve identified.
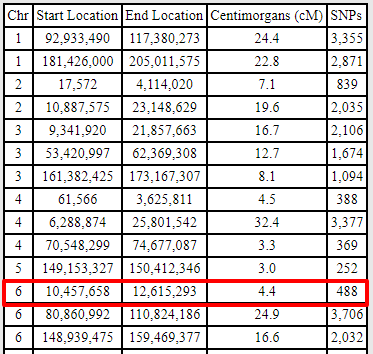
The 4.4 cM Segment
The 4.4 cM segment that remained after phasing could be a true segment created by a recombination event. When I compared Fred to Mitchell, they also share the 4.4 cM segment in addition to many other segments:
Blanche, my aunt, also shares DNA in this region with Mitchell, but it is a larger segment:
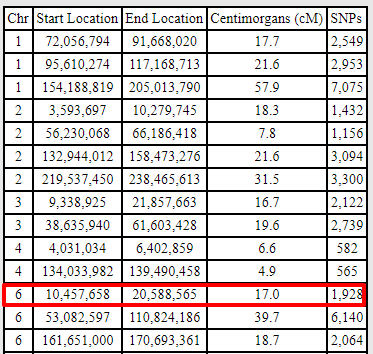
And sure enough, when I compare Blanche and Fred (remember that they are siblings), it appears they may have a recombination event between them that explains why the segment Fred shares with Mitchell is so much smaller than the segment Blanche shares with Mitchell in the same location:
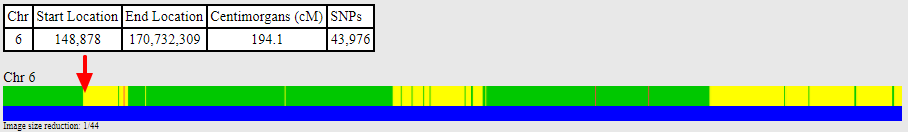
Using the full resolution view, we see a recombination event that occurred between 12,000,000 and 13,000,000:
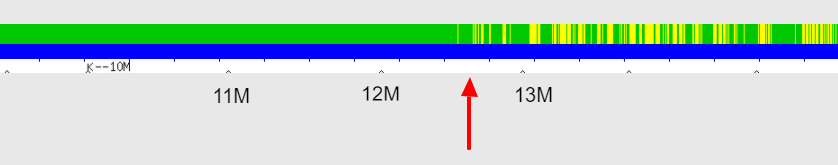
I haven’t yet visually phased this chromosome to confirm that the segment leading up to 12.5M is the paternal segment, but there’s little doubt that it will prove to be.
What is important here, however, is that I took MANY additional steps to analyze this small segment, and I have a hypothesis as to why the 4.4 cM segment exists between Fred and Mitchell. This was only made possible, however, by having another tested individual with a larger segment at that location, where I could show there was recombination that created the smaller segment. If all I had was another distant cousin that shared the same 4.4 cM segment, I would not be able to make any conclusions about the small segment.
Take-Home Lesson
There is a common misconception that sharing one or more large segments with a match means that the small segments shared with that match are real segments, and that knowing the genealogical connection with a match means that the small segments shared with that match are real segments.
However, as we saw above, phasing eliminates many false matches. Even with a phased kit, we need additional information (such as additional closely-related test-takers) to analyze any small segments that may survive phasing.
.
Excellent analysis once again.
I actively encourage people to use their phased kits for further and divergent analysis.
They might just be the little piece of information and the key to the puzzle.
Thank you!
Blaine, another great reminder. I, too, have found similar results. For several of my known, close cousins, I recorded the smaller shared segments in my spreadsheet – down to 3cM. I colored coded them light brown to warn me about them. Now, with a lot of my Triangulated Groups fairly well established beyond the paternal and maternal designation, I know that many of those small shared segments don’t fit.
A few comments however:
– The “32% don’t match a parent” type statements, particularly in the context of other evidence about false-positives, tend to lead the reader to think this means 32% false positives. In point of fact the 32% is a mix of false-negatives and false-positives. The “false” prefix is correct, but misleading, as the false-negatives are actually true Matches. I’ve studied this area in detail, using GEDmatch, and it’s my conclusion that the false-positives (above the company threshold ~7.9cM at FTDNA for example) is around 6-7%. I encourage you, and others with two parent data, to also try to separate the 32% headline-grabbing numbers into true Matches and false Matches
-As a Triangulation proponent, I find this technique culls out most of the false-positives above 7.0cM. However, I don’t think Triangulation is as accurate at lower cMs.
-Throughout you write of segments being false. Some “shared” segments, or “HIR” segments, are false. One person’s segments are true – it’s only when an algorithm compares the raw data do we get false shared segments. So “segments” in this post are assumed to mean shared segments.
Completely agree with point #1. Of course, reducing the number from 32% to 25% doesn’t make me feel too much better! 🙂
I consider your data to be among the best to evaluate point #2. You’ve probably triangulated more of your DNA than anyone in the world. I don’t have enough to evaluate either way, unfortunately.
Regarding point #3 (and correct me if I misunderstood), if the “segment” is created by weaving back and forth between chromosomes (i.e, a pseudosegment), then it isn’t really a segment.
But what can we do if we don’t have our parents DNA? Should I test my children and then use their father and me for the phasing?
There is a GREAT way to [pseudo]phase your own DNA if you have a child tested but you don’t have your parents. You can learn more here:
https://thegeneticgenealogist.com/2017/07/26/the-effect-of-phasing-on-reducing-false-distant-matches-or-phasing-a-parent-using-gedmatch/
It involves your child’s phased kit, and his/her Evil Twin.
And if my parents are long-dead and I don’t have any kids, can I test my dog? 😉
Yes, but before you test, please be aware that there can be unexpected results!
I love your cautionary advice—but small segments are not poison! They are however more likely to ambiguous and not delineate the ancestors we think—that is an important caution. However for population identification they are often quite helpful. I have many (likely very old matches) or some newer ones that point to shared ancestral populations rather than shared ancestors per se. I can make book on my Norwegian matching segments—however small– they point to Norway and nowhere else. And then there are those Southern English Colonial small segments that are not worth the bother except when separating one population from another…So love the post but poison—one man’s poison is another women’s cure… 😉
I don’t know how to get around the science that shows that most small segments are false. Finding that other people share these segments doesn’t make them any more likely to be valid, as this post discusses. But I hope you find a way to refute it!
Kelly, I completely agree. With only 1 segment being an exception in each, My and my wife’s DNA and my parents share no segments at 4cM and 700. So I feel comfortable mapping segments that are 5cM and 700 SNP’s. I feel very comfortable identifying those segments that are inherited. These numbers are not the same for all my paths. I believe that Blaine’s family tree is not reflective of any of my ancestral lines.
My paternal grandmother ancestral line was born in Poland, I have documentation going back 3 to 4 generations along these lines, and even further for my paternal grandmothers Colonial New England side of the family. There is also a division on the Polish side. One side is really from the Polish-Russian side, and the other is Austria-Poland.
It is very difficult for me to believe that one segment, however small, came from my Colonial New England side of the family and another segment came from my Polish side of the family.
Can we turn this around and quote a probability that a certan number of small segment of various sizes will show a real match? Rather than get nothing from a small segment result can we get something?
Unfortunately, no, because how are those small segments being validated? If a false small segment and a valid small segment look the same, how can we tell the difference? If the poison M&Ms in the bowl look just like regular M&Ms, would you reach in and grab some to eat?
Based on your research Do you agree with the Wikitree guidelines?
Wikitree first requires a paper trail evidence of a relationship between two DNA testers before accepting DNA evidence for the purpose of further confirming the parent/child relationships that connect them. In other words, If I have certified birth/marriage/death documents and a DNA prediction that supports the relationship, there is a significant reason to test another cousin for those relationships. They don’t consider this a positive match for relationships greater than 3rd cousins.
The guidelines require a 3rd distant cousin that is also well-documented. That distant cousin can be more distant, the same distant, or a closer distant, and that the 3rd test must triangulate with a matching size segment.
How many examples have you come across in which the documentation and the DNA are consistent but a third test will show that both the documentation and DNA service prediction was wrong for those parent/child relationships up to but not including the MRCA?
It seems that FTDNA takes the opposite position on this: their Shared Centimorgans column includes contributions from many segments less than 5 cM in length. For example, I have a FTDNA match which shows 56 Shared Centimorgans with the Longest Segment as 8 Centimorgans. Going into their chromosome browser for this match and downloading the chromosome data to Excel clearly shows that of the 18 segments contributing their reported 56 Shared Centimorgans only 2 are greater than 5 cM in length. Besides the problems this may cause in interpreting the FTDNA results, it may also affect comparisons of FTDNA results with those of other companies. In particular, your most recent Shared cM Project results show that the averages, medians, and percentile ranges for FTDNA results are significantly larger than for those of other companies. This would seem to be due in part to FTDNA including short segments in their results which are excluded by the other companies. Should users of FTDNA results be cautioned about using the averages and lower bound ranges in the Shared cM Project results to interpret their FTDNA results? Likewise, should testers with other companies understand that most of the submissions represented in the long right tails of the bar charts come from FTDNA tests?
I certainly agree with everything you say; although I have had a few dozen dna matches at Ancestry with whom I shared 6-7 cMs and found tree matches between us. My only thought is that I am 74; and perhaps my match/s were 30 years old or so, and had been through recombination 1 or 2 more times than I. Meaning they and I were not on the same playing field, and perhaps I would have had a legit cMs match with theier parent or grandparent.
A question, please. Would my 1/2 brother’s son, be my nephew or my 1/2 nephew?
If he is my 1/2 nephew, approx how many cMs would I share with him.
Many thanks!
From someone I know:
My half sister and me – 1672 across 43 – shows as “close match” on Ancestry
Half sister to my daughter 1 – 992 across 29 – shows as 1st cousin
daughter 2 – 709 across 25 – 1st cousin
Another set of relationships:
My father and his first cousin on their father’s side – 737 across 37 – show as 1st cousin
Dad’s first cousin to me – 310 across 11 – 2nd cousin
to my half sister – 456 across 24
Dad’s first cousin to my daughter 1 – 115 across 6 – 3rd cousin
daughter 2 – 52 across 2 – 4th cousin
Would it make a difference if I matched the same small segment with several people with whom I also shared longer segments? How would I calculate the probability of false matches in this case?
I have noticed you don’t monetize your blog, don’t waste your traffic, you can earn extra cash every month because
you’ve got hi quality content. If you want to know how
to make extra money, search for: Mertiso’s tips
best adsense alternative
I have checked your website and i have found some duplicate content,
that’s why you don’t rank high in google’s search results,
but there is a tool that can help you to create 100% unique content,
search for; Boorfe’s tips unlimited content
Thanks so much for this great article. I’ve been using DNApainter to work through my matches, and wasn’t sure what to do with those short matches. I have a few 3-5 cM segments that are shared with several legit matches. The legit matches sometimes have a match with me on a small segment, and a longer matched segment with a triangulated kit with a left or right “overhang”.
Anyhow, I’ll be more careful with these small segments.
This gives large opportunities. That’s really cool, though, I won’t do this right now. This year I decided to spend my money on travelling. On this website https://worldcams.tv/british-virgin-islands/jost-van-dyke/soggy-dollar-beach-bar I found a webcam from one bar on the Virgin Islands. I think it’s gonna be my destination.
Thank you for the current article. My thesis paper is also about it but it is too tough. I think I won’t manage it… So, perhaps anyone here has ordered a thesis and can answer. is edubirdie reliable? I am not so rich to make mistakes now. And I already got the fact that many of those services aren’t really reliable, sadly.
good post
I would like to add an exception. Last July before the change, I ‘grouped’ almost all of my 6-8cM at Ancestry. It turned out that 2 people had extensive trees back to 1750-1800 with a ‘brick wall’ to my extensive life long Harding ancestry. Both were from unknown Harding children who left home early, 1 across the Potomac, one to Ohio. Neither of which I had explored. The naming, dates and proximity indicated a high probability they might be connected to me. Also while they only matched 6-8cM, their first or second cousins matched 10-25 and even 50cM to me. So don’t ignore small segments as possible links to distant cousins.