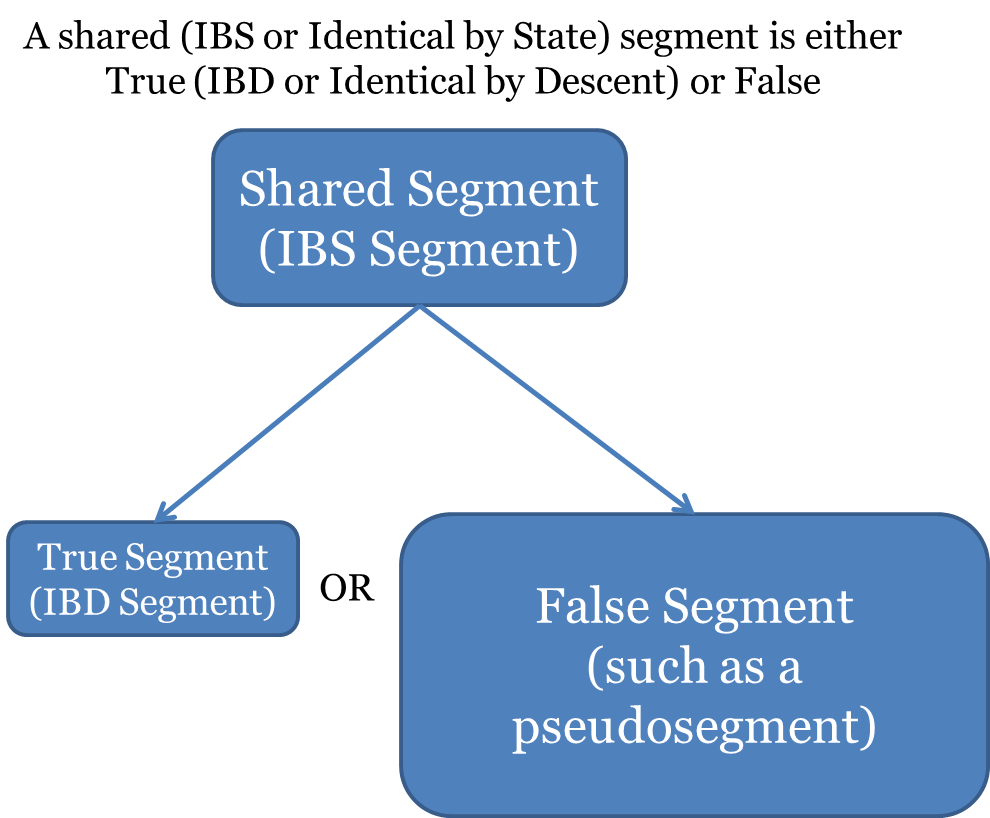
Genealogical autosomal DNA evidence relies on segments of DNA shared between two or more individuals. When they are true matching segments, they provide information about shared ancestry. One problem that genealogists are currently facing is the inability to decipher between “real” or “true” matching segments and “false” segments.
I won’t get too much into all the different terminology of “real” versus “false” here, because it isn’t important and takes away from the more important discussion. Genealogists, like patent attorneys, can be their own lexicographer, just so long as they are understood by the reader by providing a good definition. So here are my definitions for this post (and I typically use these elsewhere):
- Shared segment – also known as IBS (identical by state). IBS simply means that two segments of DNA are the same. Therefore, ALL matching segments are IBS. Accordingly, both true segments and false segments are IBS; some are are the result of shared ancestry, and some are not (see the two definitions below breaking down IBS into its two different categories).
- True or Real segment – also known as IBD (identical by descent), it is a segment of DNA shared by two or more people and inherited from a common ancestor without any intervening recombination.
- False segment – a segment of DNA shared by two or more people for a reason other than shared ancestry. False segments can result from several processes, including mutations, sequencing errors, phasing errors, lack of phasing (weaving back and forth between parental strands of DNA to create a new segment), and others. It is hypothesized (see below) that the vast majority of false segments are pseudosegments resulting from lack of phasing.
In January, I wrote “The Danger of Distant Matches,” in which I compared my list of DNA matches with the lists for my mother and my father. In that analysis, I discovered that 32% of my matches at AncestryDNA were not shared by either parent. The vast majority of those matches shared only small segments with me. Of these matches, the breakdown looked like this
- 261 share 10 cM or more (5%)
- 1784 share between 7-10 cM (35%)
- 3090 share between 6-7 cM (60%)
This was not entirely new information. In 2014, 23andMe researchers published a paper in which they showed that 67% of all phased segments smaller than 4 cM appear to be false segments, and at least 33% of 5 cM phased segments appeared to be false segments. See “Small Matching Segments – Friend or Foe?” for more information.
Avoiding False Segments
The surest, simplest way to avoid false segments is to select a segment threshold that is hypothesized to eliminate most false segments. For example, in “The Danger of Distant Matches,” I concluded that segments of 15 cM and above appear to be true segments more than 99% of the time. Segments larger than 10 cM appear to be true segments most of the time (95+%).
However, segments smaller than 10 cM should be considered suspect. This hypothesis is supported by other analyses (see “Identical by Descent” on the ISOGG Wiki).
Selecting a threshold that minimizes false segments while maximizing true segments is what the companies do with their matching thresholds. However, most company matching thresholds are set unrealistically low in order to maximize matches. Although these very low thresholds do indeed identify true segments, they also report thousands of false segments as a result.
[Note: the issue of the AGE of the segment is a related issue but won’t be addressed in detail in this post. Although a large segment is almost certainly IBD, it can still be very old. A 2015 paper by Speed and Balding showed that around 40% of 20 mB segments (which we can roughly equate to 20 cM segments) date back beyond 10 generations. Although the study only used simulated data and I am not convinced that we understand the mechanisms of recombination and inheritance to rely solely on simulated data, this is the best data we have currently. See a discussion of the Speed and Balding paper at “Identical by Descent” on the ISOGG Wiki].
Reducing False Segments With Phasing
Another way to reduce false segments is to phase a child’s DNA into the parental chromosomes. This requires tested DNA for a child and at least one parent. The Phasing tool at GEDmatch performs phasing using DNA that has been uploaded to the third-party tool:
The Phasing tool produces two files, one which is the mother’s contribution to the child (which is just 50% of the mother’s total DNA) and the other is the father’s contribution to the child (which is just 50% of the father’s total DNA). The naming pattern for these new files is thus:
- Child’s kit used to phase: A123456
- Paternal DNA kit: PA123456P1
- Maternal DNA kit: PA123456M1
The phased kits are ready for One-to-One comparisons immediately, and are ready for One-to-Many comparisons after they are processed (usually just hours or a day or two).
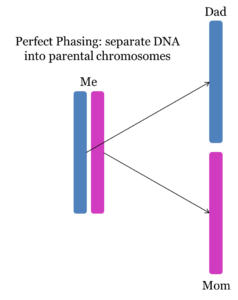
These phased kits are hypothesized to significantly reduce false segments. Specifically, since there is only a single copy of each chromosome in each phased kit (Mom’s chromosome in the maternal kit and Dad’s chromosome in the paternal kit), the matching algorithm cannot weave back and forth between mixed results to create a pseudosegment. The following illustration emphasizes this point. In the Unphased Results, the matching algorithm can form any matching segment using the results from BOTH Mom and Dad. Using the Phased Result, there can only be a match if the compared segment and the phased segment match (which they do not in this illustration):
I have tested myself and both my parents using the AncestryDNA V1 chip, and uploaded all of the results to GEDmatch. I then phased my DNA into maternal and paternal kits using BOTH of my parents (triad phasing). I then determined how many matches at GEDmatch I have with my unphased kit versus my maternal phased kit and my paternal phased kit (many others, especially Ann Turner, have performed a similar analysis).
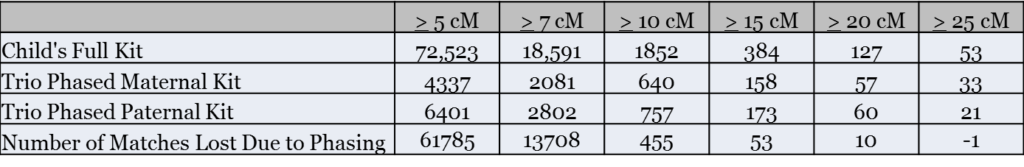
As the following table shows, phasing significantly reduced the number of distant matches I had at GEDmatch:
The biggest reduction in matches, not surprisingly, was at the 5 cM and 7 cM levels. For example, at 5 cM and greater, I lost 82% of my matches due to phasing. Even at 10 cM, I lost 25% of my matches.
In a future analysis, I’d like to review the matches I lost above 20 cM. My very preliminary hypothesis is that these were not completely lost by phasing, but were reduced to a smaller size. However, this requires additional analysis.
Phasing a Parent
In the previous experiment, I phased MY DNA using both of my parents (you can also phase with just one parent).
However, I was not able to test any of my grandparents, so I cannot phase my parents using the traditional phasing tool. I can, however, using tools at GEDmatch to phase my parent’s DNA into single chromosomes, which might significantly reduce false matching segments.
I’ll do this with a combination of the Phasing tool and the My Evil Twin Phasing tool, a Tier 1 tool available at GEDmatch.
The My Evil Twin Phasing tool produces one or two files, depending on how many parents are used. If a mother is used, the tool will create a kit containing the mother’s DNA that the child DID NOT INHERIT! Thus, this kit will contain 50% of the mother’s DNA. The same is true if the father’s DNA is used. If both parents are used, a paternal and a maternal evil twin kit are created.
Together, a phased maternal kit and an evil twin maternal kit should contain (approximately) 100% of the mother’s DNA, separated into single chromosomes!
However, although these kits are separated into single chromosomes, they are not perfectly phased into the mother’s contribution from her mother and her contribution from her father. Instead, these kits are “imperfectly phased”; separated into single chromosomes which are a mix of the mother’s mother and father. The following two diagrams illustrate the difference between “perfect phasing” and “imperfect phasing”:
I then determined how many matches at GEDmatch each parent has at GEDmatch using each of the following kits:
- Parental unphased kit;
- Parental phased kit using the Phasing Tool (“Perfect Phasing”); and
- Parental phased kit using the My Evil Twin Phasing Tool (“Imperfect Phasing”).
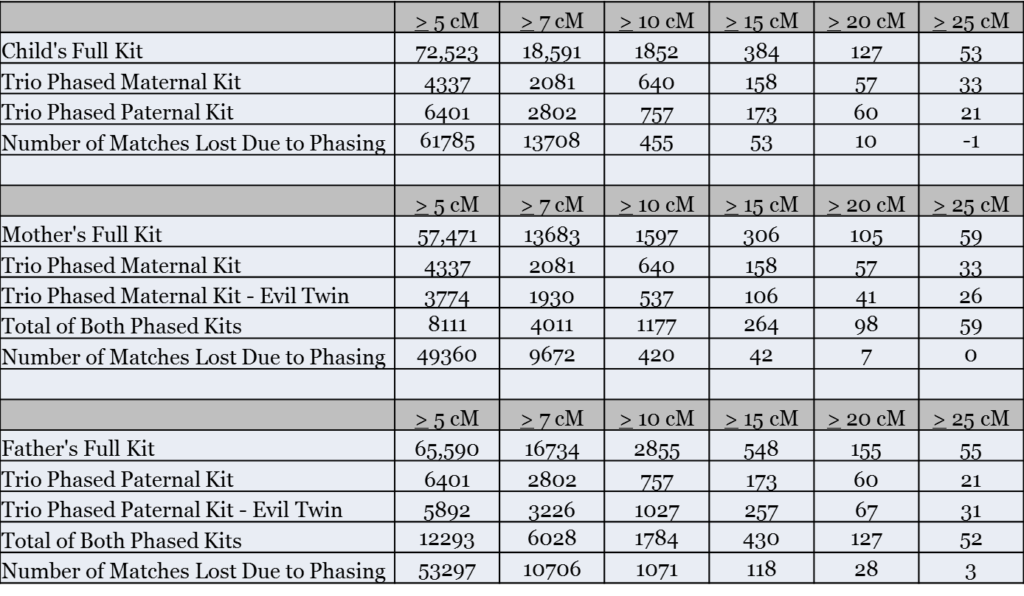
As the following table shows, phasing significantly reduced the number of distant matches
Once again, the greatest reduction in matches was below 10 cM. At 5 cM and greater my mother lost 86% of her matches, and my father lost 81% of his matches. At 10 cM and greater, my mother lost 26% of her matches and my father lost 38% of his matches.
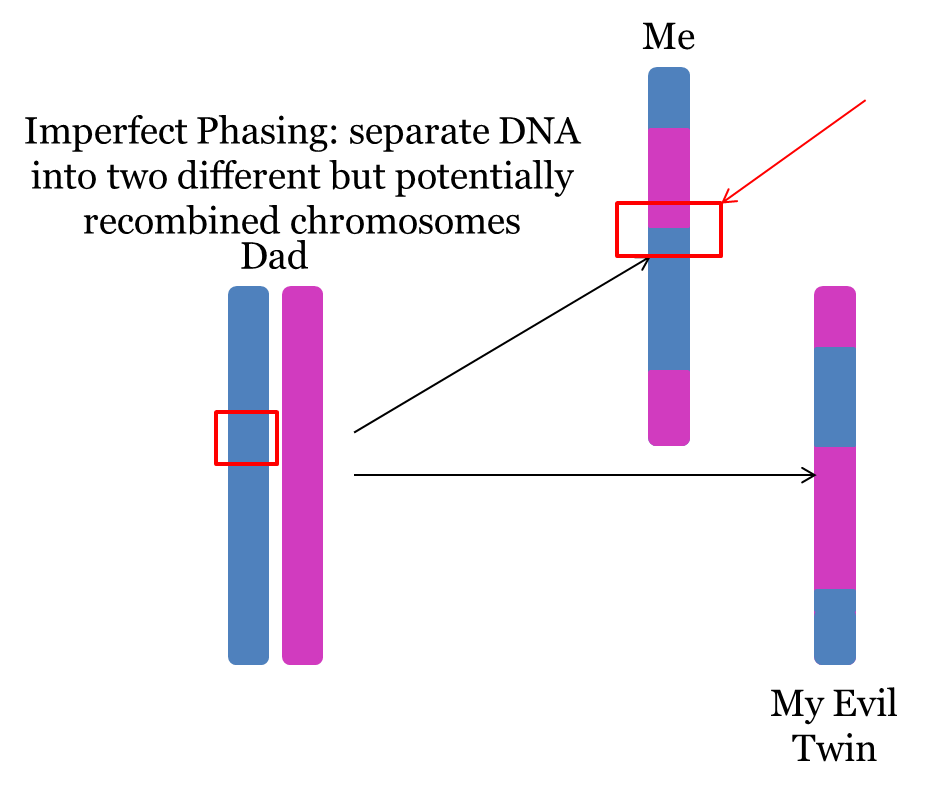
Notably, Imperfect Phasing WILL destroy some true large segments. As shown in the following diagram, a segment that matches the blue (paternal grandfather’s) chromosome in the red box may be shortened or lost due to recombination in the middle of that segment. Phasing will not recover that segment:
Accordingly, we must be careful about making unsupported conclusions using these imperfectly phased parental chromosomes.
Why Phase Parental DNA?
We might create parental phased kits in order to work with smaller segments with increased confidence in those segments. For example, we may have much greater confidence in the segments 10 cM and larger as a result of this phasing (keeping in mind the age issue described above).
There may be other interesting uses for these kits, and I’d love to hear your thoughts!

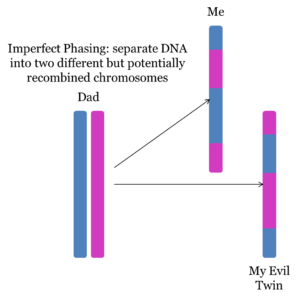
Is there any way of using kits of aunts or uncles to create a pseudo parent? Both my parents are deceased, but I tested two uncles on my paternal side and two aunts on my maternal side. Not having parental results is limiting and I’d like to figure out a way to use the data I have available.
Thanks
Lynda
An interesting study, Blaine. It is also surprising that above 15cM there were 14% of the child matches not in a parent. For several years now, despite calls for data, “no one” has reported any non-IBD segments over 15cM. So I’m thinking this analysis is more about the GEDmatch algorithms, or some other parameter. Have you looked at some of these “non-matches” in the graphics view of the GEDmatch browser? GEDmatch allows some mis-matching SNPs, but I always wonder what we really get in a phased kit… I use an M1 phased kit, and haven’s seen anything unusual.
14% above 15cM just doesn’t seem right… I think we would have determined some of those segments other ways over the past 7 years.
If I’m calculating your percentages correctly, you lost about 25% of your >= 10 cM matches, 14% of your >=15 cM matches and 8% of your >=20 cM matches. My numbers, and those of my 2 sibs are running roughly the same as yours: 25% /11% /3% for me, 24% /17% /2% for my sister, and 23% /14% /5.5% for my brother.
That said, based on Jim’s comment (above), I went back and did a 1-1 match with each of my own “lost” phased matches for the >= 15.0 cM group. Each “lost” matches DID in fact match either mom or dad. My parents just had smaller segments. For roughly 25% of the “lost” matches, my parent’s segment was in the 14.5 cM – 14.9 cM range, but in other cases, their matching segment was as much as 3.6 cM smaller than mine.
Can I use any of this with a half sibling to help find my mystery father. Half sibling and I have the same mother but not the same father. We do have DNA for her father.
Blaine, this concept blew my mind!
My mum passed away in 2009, and not long before that I became aware that her dad (my granddad) was not her biological dad, and she died not knowing this. She was one of 8 children, and was the only one with a different dad, so testing my aunts and uncles won’t assist me here.
I have my siblings and dad (and his mum) tested, and I would love to learn who my bio granddad was.
Using your model above, I have just phased my kit and my siblings kits. In theory, if I was to use my phased maternal kit in the ‘My Evil Twin Phasing tool’, would running the ‘one to many’ report on my phased maternal kit and maternal evil twin kit give me some DNA matches of my mum?
I am at an absolute brick wall here in my research, and am cautious that this looks like a mirage to a lost explorer in the desert at the moment. I’d love your feedback here as to whether I could get some reliable matches for mum this way.
By the way, you can use the Gedmatch phasing tool to phase a parent in one step. Just enter the parent’s kit as the “child” and the child’s kit as the sex-appropriate parent. All the same caveats apply as using the child’s phased kit plus the evil twin kit.