I’ve always known that I have weird mtDNA. This morning, I learned that it is so weird that it has helped identify a new branch of the mtDNA Haplogroup A family tree.
Haplogroup A
When I first received the results of an mtDNA test ten years ago, I was shocked to learn that my mtDNA belonged to Haplogroup A, one of the well-known Native American haplogroups. I knew that my ancestors on my maternal line were British missionaries to Roatan, Bay Islands, Honduras, traceable back to about the 1820’s, and so I was expecting haplgroup H or another traditionally European mtDNA haplogroup.

My mtDNA Line – 5 Generations in Photographs
Not surprisingly, I had no close matches in the Family Tree DNA database. And when I tried to research my haplogroup, I had a handful of control region mutations that no one else in the database or the academic literature seems to have.
My mtDNA appeared to be an outlier. Most vexing of all was the fact that every A2 sequence I looked at had a mutation at position 16111, a mutation that I was lacking. I would often find sequences that were very close to mine, but invariably they would have a mutation at 16111 while I would not.
In 2013, I finally upgraded to a full mitochondrial DNA sequence from Family Tree DNA. With these results I finally had matches, all at a genetic distance of 1 or more (which is why I didn’t see them at the HVR1/HVR2 level). I was lucky enough to determine that we were all related and all had roots in the Bay Islands. However, we didn’t learn anything new about our particular branch of the A2 haplogroup.
I did made sure that I made my full mitochondrial DNA sequence available to Genbank, which is a database of public DNA sequences from all over the world including many hundreds of mtDNA sequences from Family Tree DNA. My “Accession Number” for Genbank was KJ018969, and my mtDNA was available to anyone for research.
Further Down the Tree – Haplogroup A2w1
Last year, Family Tree DNA changed my haplogroup from A2 to A2w:
This was based in part on a 2011 paper entitled “Large scale mitochondrial sequencing in Mexican Americans suggests a reappraisal of Native American origins” which identified several new subclades of A2. PhyloTree, which is the most detailed phylogenetic tree of global human mtDNA, soon added A2w to the tree, followed by yet another subclade A2w1:
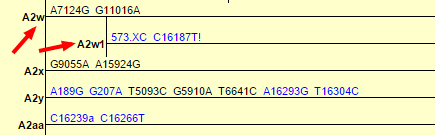
This morning, while looking over my results from Tribecode, I saw that their test had also categorized me as subclade A2w1. Determined to learn the latest about A2w1, I did a quick search of both Google and the academic databases (Google Scholar, PubMed, others) to find anything I could about the subclade.
As a result of the search, I uncovered a paper from February of 2015 that included another surprise for my mtDNA!
Studying the Mayans – Haplogroup A2w1b
Earlier this year, a group in Spain published a paper that explored the genetic variability of the Maya and Ladinos from Guatemala, entitled “Genomic insights on the ethno-history of the Maya and the ‘Ladinos’ from Guatemala.” During the course of the study, the researchers obtained numerous mtDNA sequences with roots in Mesoamerica, including 32 full genome sequences within haplogroup A2w (a large number of them analyzed within The 1000 Genome Project in a Colombian sample set). Using these sequences, the authors identified five new subclades within A2w, namely A2w1a and A2w1b (which are subclades of the existing A2w1), and the new sister clades A2w2, A2w3, and A2w4.
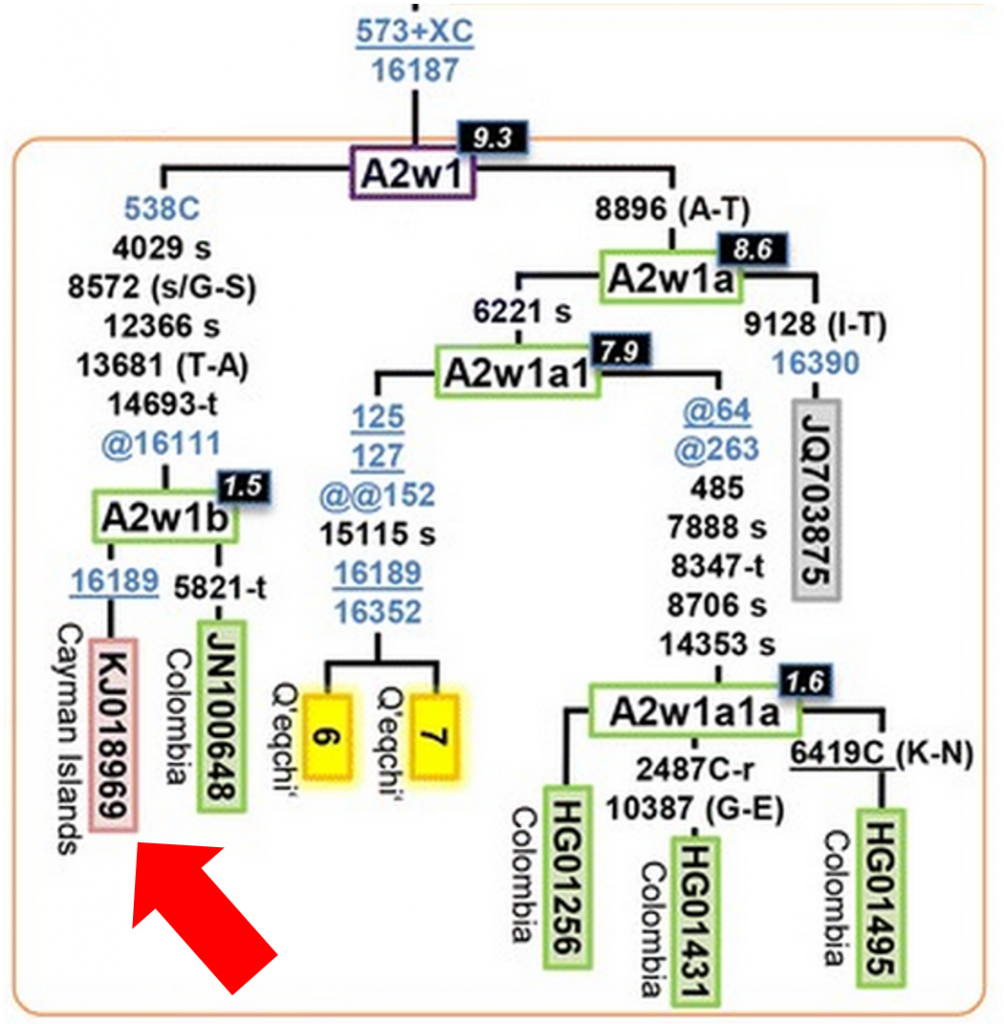
Looking at figure 5 of the paper, I zoomed in on A2w1, the clade where I knew I likely fit:
And there I saw a familiar number and location, sequence KJ018969 from the Cayman Islands, my mtDNA sequence.
Together with JN100648 (another full sequence from FTDNA, with roots in Columbia), my mtDNA sequence had helped define the new A2w1b subclade. And even the two of us differ by one new mutation each and coalesce about 1,500 years ago, suggesting that we may one day form our own subclades (maybe I’m A2w1b1?).
There’s so much interesting information in this figure. For example, this confirms what I’ve always suspected, that my lack of the 16111T mutation is due to a back mutation (denoted by “@16111”) within my subclade. Additionally, A2w1b appears to have split from A2w1a approximately 9,300 years ago, so this is a very old subclade. We also have a long list of mutations separating us from A2w1b, including 1 control region mutation (in blue), 5 mutations outside the control region (in black), and one back mutation.
Conclusions
We are citizen scientists. Genetic genealogists have contributed so much to the understanding of the Y-DNA and mtDNA haplogroup trees. From the ISOGG Y-DNA Haplogroup Tree to a new root of the Y-DNA tree to thousands of mtDNA Genbank submissions, there is a wealth of public information available to benefit all science, paid for courtesy of thousands of genetic genealogists.
Submit your full mitochondrial genome sequence to Genbank (of course being mindful of the potential for disclosure of mitochondrial conditions/disease). You never know who it might benefit or how it might be used for science.
Wonderful, Blaine, congratulations! I’m excited to learn of this paper on Mayan and Guatemalan population genetics, as the history of the Maya in Guatemala was the subject of my undergraduate studies at Friends World College, Fascinating to see all we’re learning now as more peoples are sampled and the phylogeny grows ever more detailed. I never would have expected the genetic connection between Guatemalan and Colombian peoples. Your mtDNA has traveled a long way!
It really has traveled a long way! And what a rich heritage it has, I’m very lucky to be able to know more about my mtDNA line thanks to these types of excellent studies.
Whee! It is exciting to see how quickly the science can advance with input from genetic genealogists. I looked back at the custom report I wrote for you a year and a half ago, when you were still A2w. I predicted a new subclade would be forthcoming, and sure enough, you became A2w1 with the next version of Phylotree. And now you are A2w1b! Phylotree typically requires three distinct haplotypes to add a new subclade, so you may not make it into the next version. However, they do make some exceptions for labels that have appeared in the technical literature, especially if it’s clear from geography that the sequences are not closely related genealogically.
I hope your example will encourage more people to upload their full mitochondrial sequences to GenBank. Every sequence (not just “exotic” ones like yours) can make a contribution to understanding when mutations occur and how the tree develops. Ian Logan has instructions on how to do this on his website http://www.ianlogan.co.uk/Submission.htm.
You called it Ann! My mtDNA stuck out like a sore thumb, it’s so much fun to see it fall into line in the family tree!
I agree, it’s going to be a long time before the clades below A2 are well-defined. Luckily there’s been a continuing interest in understanding the genetic heritage of Central America and the Caribbean, so I expect there will continue to be testing that elucidates these clades. I’ll just sit back and watch!
I am glad to know that I am not the only one that has weird mtDNA. When I was tested, I had NO matches, so I thought that this must happen to everyone and it is rare to have matches. Nope! Not the case as I learned from talking to others, and I seem to be the oddball. I am T2b7a1, and have an extra mutation that no one else who has tested seems to have. If you take away that extra mutation, there are some matches, but not a ton. I don’t know what to do to even make this information useful for me!
Hi I just read your article and wanted to chime in as I took a test with FamilyTreeDNA and was also found to be of haplogroup A. Now, I did not take the full sequence test but the MTDNAplus one which did little to narrow down my haplogroup. I contacted a person named Roberta Estes as she was doing a study on A4 haplogroups and she seems to think I belong to some sort of A4 subgroup. One of the reasons is that I lack the 16111 mutation as well. I have since upgraded to the full sequence to find out what the actual group is. I am from Colombia originally (both parents are Colombian, Valle Del Cauca region) so it should be interesting to see what I get.
This is one of the best blog posts re mtDNA that I have seen, and the hyperlink to phylotree with its extensive tree structure is excellent !!!
Thanks once again !!!
This is very interesting research papers. I believe such subjects are important and they need to be spread. I myself can’t handle my works properly. Deadlines are soon and I believe I won’t hit them. So, I want to pay for research papers on uk.edusson.com. What are your thoughts on it? Do you believe such services? I read lots of positive feedback on them.