If you aren’t already a member of the coolest Facebook group ever, Genetic Genealogy Tips & Techniques, you really should be! We have a friendly and engaging environment, and everyone learns something new every day!
This post is meant to answer a question or issue that is raised almost daily in the group, and that is the issue of small shared DNA segments. Although these small segments are alluring, they are the mythological sirens of the genealogical world!
Small Segments Executive Summary
Here’s a bite-sized summary of the content below:
- Many to most small segments (at least 7 cM and smaller) are FALSE, meaning they are NOT actually shared by the two matches, and therefore do NOT indicate shared ancestry;
- This is supported by a 2014 paper by 23andMe scientists showing that at least 33% of 5 cM phased DNA segments are false-positive (and it’s much worse for unphased segments or segments smaller than 5 cM);
- This is further supported by evidence that anywhere from 20-35% of distant matches at a testing company are not shared with either tested parent;
- This is further supported by evidence that phasing your DNA with two tested parents significantly reduces the number of matches below 10 cM (with proportionally more matches reduced as the segment size gets smaller);
- There is currently no evidence that triangulating segments or finding a paper trail provides a mechanism for distinguishing between false segments and valid segments;
- Since we can’t tell the difference between false small segments and valid small segments, we must avoid these small segments to avoid poisoning our genealogical conclusions with false data; and
- Beware any research or conclusion that uses these small segments without specifically addressing the issues that are known – based on all the scientific research and evidence gathered to date – to surround small segments.
If you’re interested in learning more, keep reading!
Small Segments In Detail
One of the most common questions in the group has to do with small segments. There’s no exact definition of “small” when it comes to small segments, but many of us define them as being a single segment of DNA of 7 cM or smaller. Others use 5 cM or smaller, while others use 10 cM or smaller. Personally, I consider segments of 7 cM or less to be “small,” although when I’m being very conservative I use a definition of 10 cM or smaller.
The issue of small segments often arises due to GEDmatch, where we can mine our matches for these small segments. Although GEDmatch sets most of its matching thresholds at 7 cM (meaning you have to share a segment of at least 7 cM with a match to be considered “a match”), we can sometimes lower that threshold.
For example, here’s a One-to-One comparison of two people that I have no reason to believe are related:
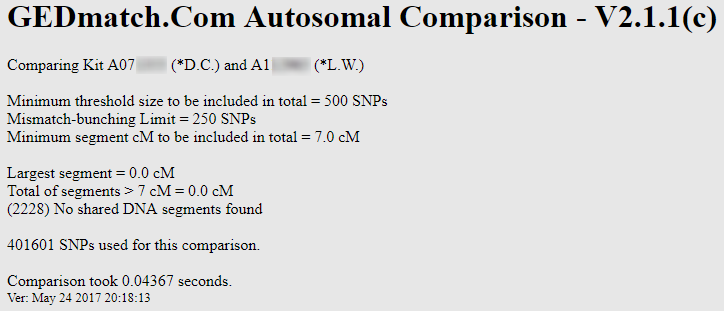
Sure enough, no DNA shared using the default 7 cM threshold. But what happens when I lower the threshold to say 2 cM (and the SNP threshold to 300 SNPs per segment):
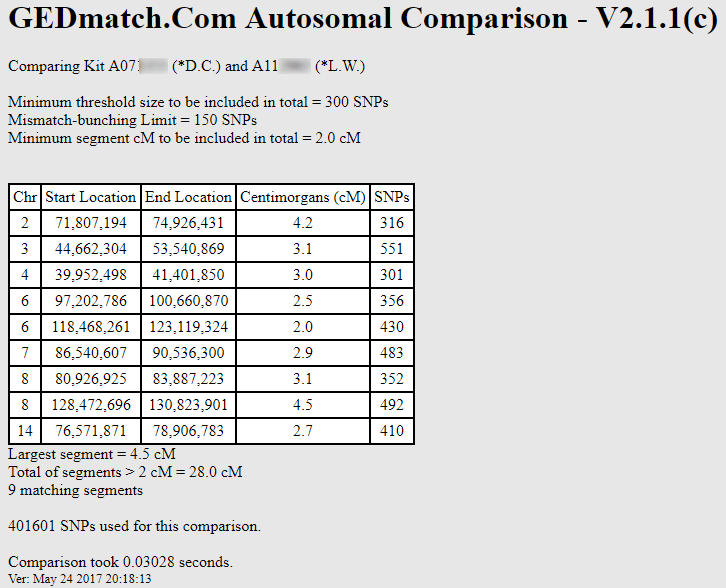
Now they share 9 segments! If I continue to lower the cM and SNP thresholds, I can see even more shared segments. Try it for yourself: my GEDmatch Kit # is A812216. How many segments do we share when you lower the thresholds? It’s almost impossible NOT to find shared segments if you lower the SNP and cM thresholds!
Does this mean that D.C. and L.W. are related? Not from this data, no. We can make absolutely NO conclusion with this data, since we don’t know whether any of these shared segments are valid (not to mention that they’d be ancient segments, but let’s take one issue at a time).
Here’s our hypothesis about small segments, based on ALL the available science:
Many small segments are FALSE, meaning they are not actually shared by the two matches, and therefore do not indicate shared ancestry.
Why do I say that? Well, let’s look at this a bit more. And below there are a bunch of additional posts if you’re interested in reading more (which, as a genetic genealogist, you should be!).
SCIENCE!
Some of the best science we have on this subject came from researchers at 23andMe, from a paper they published in 2014. In this paper, the researchers found that more than 67% of phased DNA segments shorter than 4 cM are false-positive segments! At least 60% of 4cM phased DNA segments were false-positive, and at least 33% of 5 cM phased DNA segments were false-positive. The paper is available online for free (http://mbe.oxfordjournals.org/content/31/8/2212).
I note that much more research is needed in this area. However, this paper from 23andMe is currently the best peer-reviewed research that deals with these small segments directly. In the meantime, many of us in the genealogical community have done studies ourselves to look at the phenomenon of small segments, and we’ve found some concerning issues that lead us to ignore these segments. For example, I did the following studies and published the results here on the blog:
- I showed that as much as 30% of my matches at AncestryDNA do not match either parent (and I’ve found similar results at Family Tree DNA). This suggests that there are many false positives and/or false negatives, and I hypothesize that this is most likely due to false small segments (see “The Danger of Distant Matches“).
- I showed that phasing DNA significantly reduces the number of matches or matching segments we have, which suggests that many unphased matching segments are false (see “The Effect of Phasing on Reducing False Distant Matches (Or, Phasing a Parent Using GEDmatch)“).
Many people have repeated these studies (see the links below).
Unfortunately, despite the published research and these duplicated studies, many people continue to use small segments.
Pseudosegments
Often, these small segments are “pseudosegments,” created by the matching algorithm weaving back and forth between unphased parental chromosomes, as shown in the following diagram. If the DNA were phased (“Phased Results”), then there would be no match to the Compared Segment.
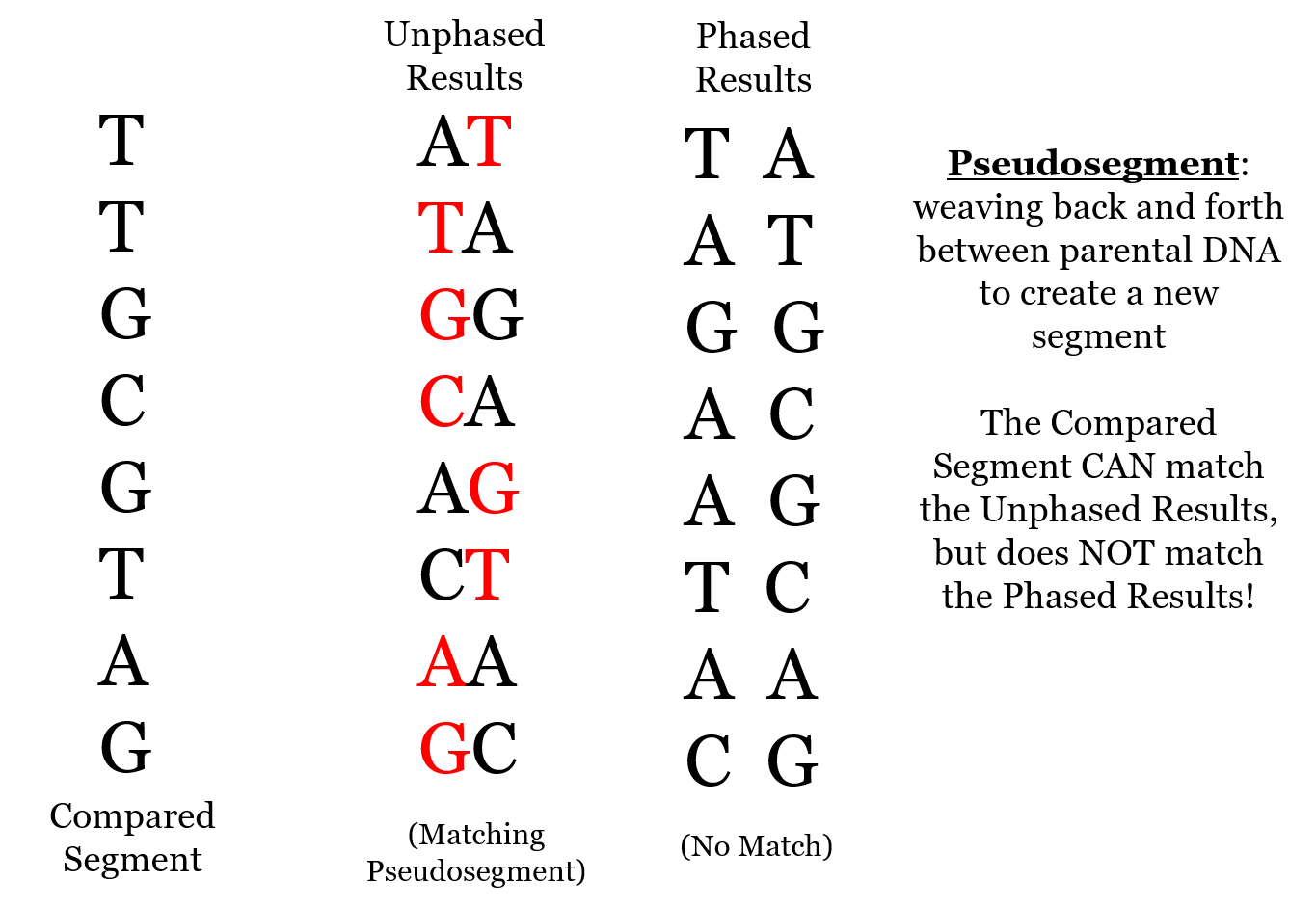
Phasing significantly reduces false matching, although as the 23andMe paper shows, even many phased segments appear to be false matches.
Poison Segments
I commonly refer to small segments as being “poison,” since they potentially poison our genealogical research. If we know that a significant percentage of small segments are false (and it is likely MOST small segments when we’re talking about small, unphased segments below 7 cM), and we don’t have any discernible way of deciphering between valid small segments and false small segments, we must consider all small segments to be poison.
To illustrate this, I use a “poison M&M” analogy:
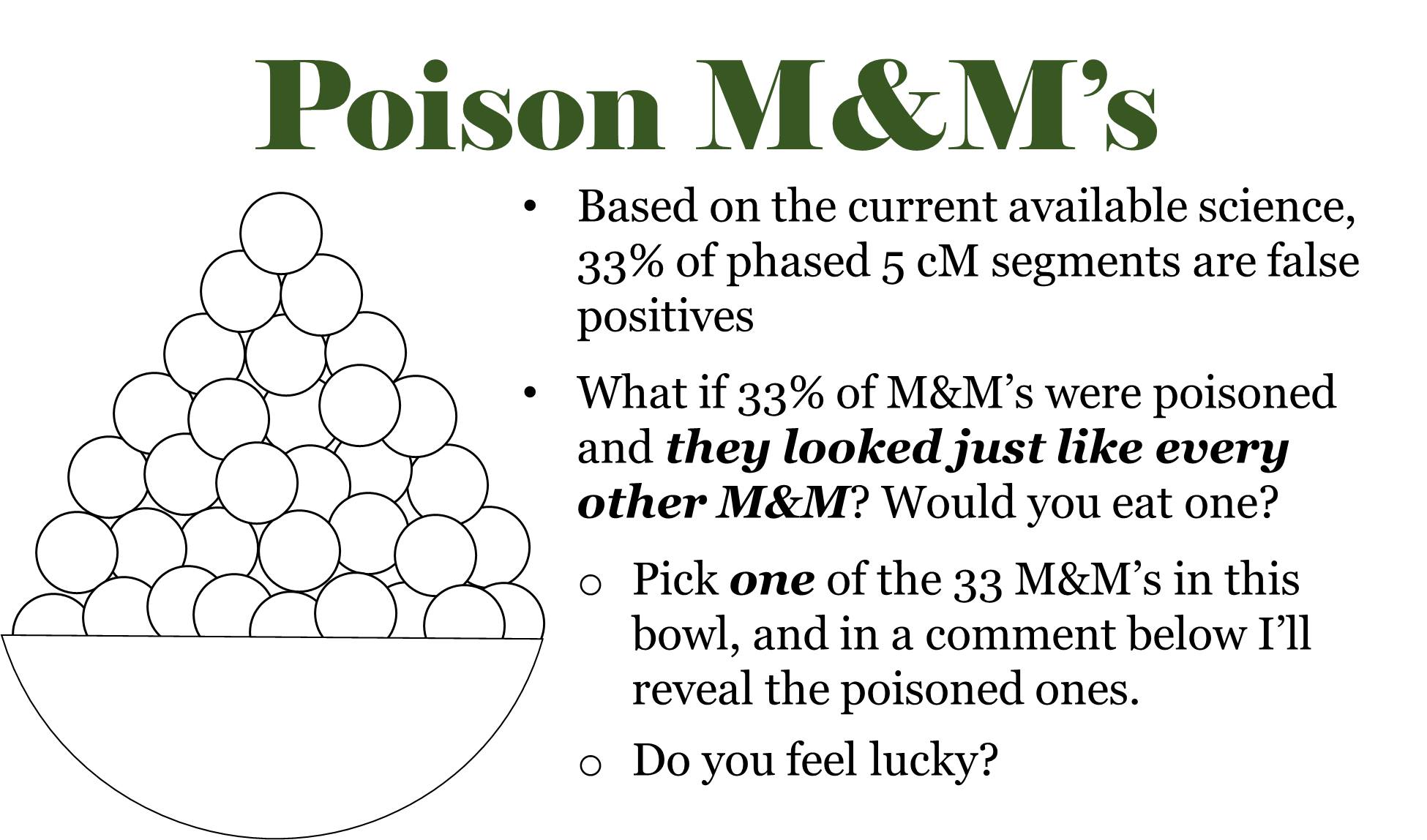
If you’ve picked one of the M&Ms in the bowl, click HERE to see if you were poisoned!
Unfortunately, false segments are not labeled red; they “look” just like every other segment.
But, But, I Have a Paper Trail!
There have been many – very good – suggestions about how to filter out false segments and use the valid small segments.
The Verified Paper Trail – For example, one suggestion is that a small segment is more likely to be real if there’s a verified paper trail showing a common ancestor. Unfortunately, there’s no support for this hypothesis. Additionally, since these small segments are likely to be many generations and many hundreds of years old, having a recent verified paper trail isn’t very meaningful. Indeed, it becomes increasing difficult to reliably show that two people aren’t related once you go back many generations and many hundreds of years old, especially since most trees get significant holes once you go back a few generations.
Shared with a Close Relative – Another suggestion is that a small segment is more likely to be real if it is shared with a close relative. Unfortunately, there’s no support for this hypothesis. It’s entirely possible that the segment is a false segment for your relative for the same reason(s) it’s a false segment for you. Or, that it might be a valid segment for you and for your cousin, but it’s a pseudosegment for your shared match. One enormous problem with small segments is that it can be a pseudosegment for anyone that reportedly shares the segment.
Triangulation (Sharing with Distant Relatives) – Another suggestion is that a small segment is more likely to be real if it is triangulated with two or more distant matches. Once again, there’s no support for this hypothesis. Ann Turner recently showed in an experiment that triangulation increases the likelihood that a segment will survive phasing, although it’s impossible to know without phasing which of these triangulated segments will survive phasing. Additionally, the 23andMe paper showed that even phased segments can be false.
The Future
I do believe that there is hope for small segments. New tests and new methodologies may be able to identify which small segments are valid and which are false. Phasing, for example, is very good at eliminating many false segments, which helps ‘concentrate’ valid segments.
In the meantime, we must be careful to avoid small segments. If you see someone using small segments without specifically addressing the issues raised herein, be extremely cautious with their research and their conclusions.
Reading More
Here are some links to read much, much more about the dangers of small segments:
- Small Matching Segments – Friend or Foe? (2 December 2014)
- Small Matching Segments – Examining Hypotheses (8 December 2014)
- GUEST POST – What a Difference a Phase Makes (30 March 2015)
- The Danger of Distant Matches (6 January 2017)
- The Effect of Phasing on Reducing False Distant Matches (Or, Phasing a Parent Using GEDmatch) (26 July 2017)
- The Folly of Using Small Segments as Proof in Genealogical Research (CeCe Moore, 3 December 2014)
- Tracking DNA segments through time and space (Debbie Kennett, 26 April 2015)
- When is a Match a False Positive? (Ann Raymont, July 2016)
- Comparing match tallies for family members with Family Tree DNA’s Family Finder test (Debbie Kennett, 29 July 2017)
- Comparing parent and child matches at AncestryDNA (Debbie Kennett, 6 August 2017)
- ISOGG Wiki, Identical By Descent
Blaine,
I really appreciate this post. For the past couple of weeks, I have been reading everything I can on the validity of small segments in relation to a geographical study I was working on. It had become abundantly clear that I could not rely on small segments. This post CLEARLY outlined the reasons why. Thanks for this.
Hi Blaine,
I really appreciate all your articles and this one is no exception. I realise that my question which follows is not on the topic of this article. However,what can one do in terms of phasing if both parents are deceased.
Thanks
Hi Claire – test your children, if you have any. Then you can pseudophase – https://thegeneticgenealogist.com/2017/07/26/the-effect-of-phasing-on-reducing-false-distant-matches-or-phasing-a-parent-using-gedmatch/
Blaine,
thank you for a great article, explained simply and clearly
dermot
Hi Blaine,
Does it mean anything if the cm’s are under 7, but the SNP’s are extremely high (4000/5000)? I have noticed this a lot on Chr. #6. Thank you!
Hi Cathy,
Check out these articles for more information about those segments on chromosome 6:
http://www.jogg.info/pages/62/files/SatiableCuriosity.pdf
https://isogg.org/wiki/Identical_by_descent (see the “Excess IBD sharing” section)
Regards,
Blaine
Great information! Thank you Blaine! I recall that you’ve also covered this in an excellent Legacy Family Tree Webinar. I’ve since started using 10cm as my smallest segment. However, I recently had a personal “small match” case where paper trails point to my being related to a person, but he didn’t show up as a shared match on Ancestry DNA for me nor my 4 siblings. The shared ancestor would be 8 generations back for me and 9 generations, for him. I would think that it would be not likely to have shared DNA that far back. Three of us siblings uploaded to GEDmatch as did this possible distant cousin. My siblings had 0 shared segments, even lowering the threshold to 2 CM. But I have a 5.6 cm a 4.4 a 3.7 a 3 and a bunch of 2s. I realize this is exactly the kind of ‘Poison” you are referring to, but I suppose this to be a good example of DNA not to be taken too seriously.
So just to clarify, if two people have the same surname, appear to share paper trail way back but only match when threshold is reduced to 6cm I still have to assume no DNA match? I cannot assume a very distant match?
OK, good article overall, but I’m confused about the M&M analogy. Picking one out of 33 is only 3%, not 33%. Maybe show a bowl of 100 where 33 are poison… or a bowl of 10 where 3 are poisoned… although who puts out a bowl of 10 M&M’s?
Hi there,
I tested with 23andme and ancestry- my dad is on ancestry but not 23andme and my mother refuses to do it.
I have a distant relative on both services who I share 15cM with (so a certain match) and our trees both link us to a man called Thomas Pettitt with his mistress Jemima Wallis (married name Reid).
We match on chr.2 in a long segment.
Thomas was married and has descendants through this marriage- the distant relative matches with those descendants on a stretch which includes our matching segment but I DONT match with them (other than one).
However, her match with them overlaps with my match with her- it starts earlier on the shared Chromosome and ends earlier whereas the length continues in her match with me on the same chromosome (ours like I said is a whole 15cM).
I checked on gedmatch progressively lowering the amount until me and all these shared cousins matched at 4cM which yes I’m aware is not good enough to be a match but because it overlaps with DNA which we know I share with a proven cousin and our paper is clear surely this would count as a true positive DNA match but where I have recombined more and thus have less trace of this distant ancestor?
Thomas was 7 generations ago and neither me nor these distant relatives have any other shared ancestry in this region.
It seems that if we’d dismiss this dna we would have to also dismiss her link with them because when you remove what insignificant percent I share with them it would remove a chunk out of what she has shared with them, lol.
So wouldn’t you say this is likely genuine?
By the way, if you are going to continue this research then you may be interested in a great solution with pautang online philippines. This happens very quickly and you absolutely do not need to go anywhere.
It seems that it makes sense, I had been considering this for a while,
and you knowI made exactly the exact decisions. I remember
the times of the last year at uni, that was a whole mess, daily and without the conclusion. The only
thing I decided to do would be to order a number of
my academic missions at essay services. Well, guess what a relief it had been. I struggled a few weeks with my essays, and this service https://essayhub.com just saved me!!!
I graduated with good grades, and even now I am so grateful for the service they
give me back afterward.
Thank you Blaine Bettinger for a great article showing how small segments can mislead research. You included an example of a random person who matched you on multiple small segments to a total of 28cMs. Do you know if any study has shown what the average amount of cMs two randomly selected people might be expected to share as a total of small segments?
Tracy L. Meyers it has also been “my experience” that we are just now tipping the iceberg so to speak where family members at distances past eight generations “thru-Lines” are actually testing and we can make the connections based on hits with an Uncle, and two Aunts and first cousins – 6cM -7 cM hit on three or four of them along thru Lines has been very very helpful in identifying and leading to matching family Lines that for years “traditional historians” have denied were if the same or would have a common ancestor. It is frustrations that Ancestry would begin to do this now when so much progress has just begun to come to fruition – I have spent five years and a lot of money in this product to just have it stripped away because of someone else’s opinion.
To be fair, it isn’t an opinion. The problems with small segments have been identified by scientists and published in peer-reviewed scientific journals.
This is ok an article but you fail to take into account endogamous populations. I presume your talking about general populations here.
I descend from Ashkenazic Jewish Mexican and Sephardic Jewish mix I’ve got tons of distant cousins from these groups as we all descended from a closed population.
Sorry ok as an article I meant typo.
Endogamy doesn’t make small segments any more likely to be valid, unfortunately.
The article says to compare the kit A812216 with your own on GEDMatch, yet when you try it says kit2 A812216 is not found in the database.
All small segments are not the same. An independent small segment of 7cM or less may be considered true or false but a small segment that is part of a collection of segments is more likely to be true. Bayes Law should be applied. Clearly a 2nd cousin that is matched at 60 cM should give the same result as his brother at 120 cM. In the first case Ancestry ignored several segments less than 6 cM. So clearly a segment of 6 cM should not be ignored. MyHeritage matching gives the right result. Both brothers match at 120 cM. I can cite many examples.
Respectfully.
This is fantastic information! Thank you so much, Blaine! I believe you mentioned this in your fantastic Legacy Family Tree Webinar. Since then, I’ve been utilizing a 10cm segment as my smallest segment. However, I recently had a personal “small match” instance, in which paper trails indicated that I was related to a person, but he did not appear as a shared match on Ancestry DNA for me or my four siblings. For me, the shared ancestor would be 8 generations back, while for him, it would be 9 generations back. I don’t think there’s any way they could have exchanged DNA that long back. Three of our siblings, as well as this possible distant relative, registered on GEDmatch. My siblings and I had no common segments, even when the threshold was lowered to 2 CM. However, I have a 5.6 cm, 4.4 cm, 3.7 cm, 3 cm, and a bunch of 2s. I realize this is the type of “poison” you’re talking about, but I think this is a good example of DNA not being taken too seriously.