I don’t match my mother’s mtDNA test results.
How is that possible?
Last fall during the sales at Family Tree DNA, I ordered an mtDNA test for my mother. Now, usually there isn’t a need to test your mother’s mtDNA if you’ve tested your own, and vice versa. You should have the same mtDNA results, and in almost all cases you will.
However, I decided to test my mother because I have a mutation that puts me at an mtDNA genetic distance of 1 relative to matches that are likely related about 200 years ago in the Caymans where my mtDNA came from. That mutation always bugged me. Although it certainly isn’t unusual to have a GD=1 at 200 years, my maternal line is a work in progress so I want as much information as possible. So I decided to “walk back” the mtDNA line as far as I could. Although my mother was only one generation further back, it couldn’t hurt to have her mtDNA sequenced just in case it could shed light on this mutation (and, frankly, the sale was too good to pass up!).
When I received her test results and logged in, I was surprised to learn that I am not in my mother’s match list, and she is not in mine! Since we both have full mtDNA sequences, that means that we have a genetic distance of FOUR or greater! Four mutations in a single generation? How is that possible? As it turns out, it may not be as difficult as we think.
A Poly-Cytosine Stretch
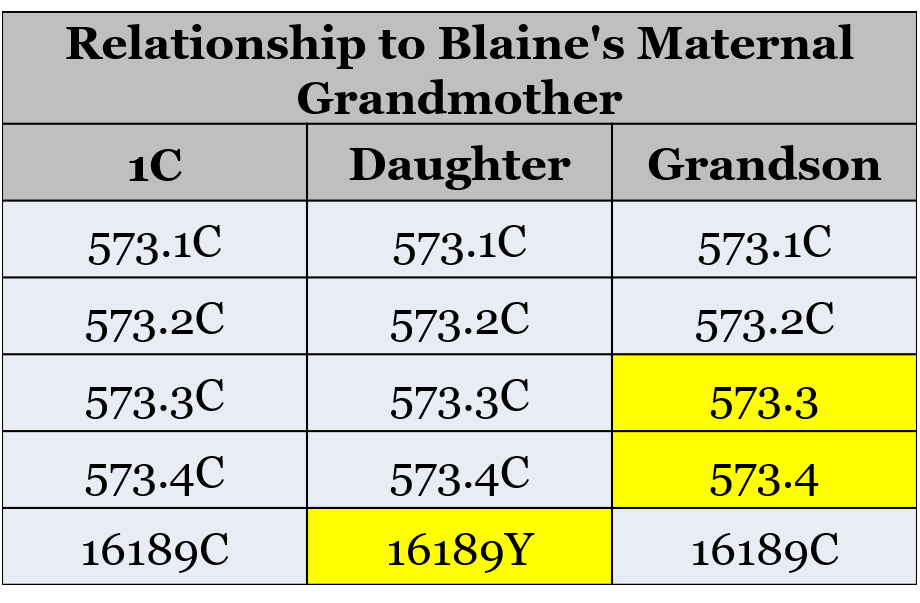
When I compared my results to my Mom’s results, I see the four differences that prevent us from being shown as matches. We match exactly, with the exception of the differences shown in this table:
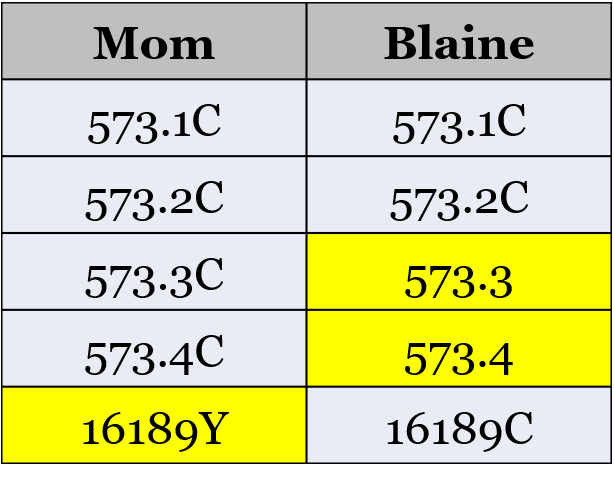
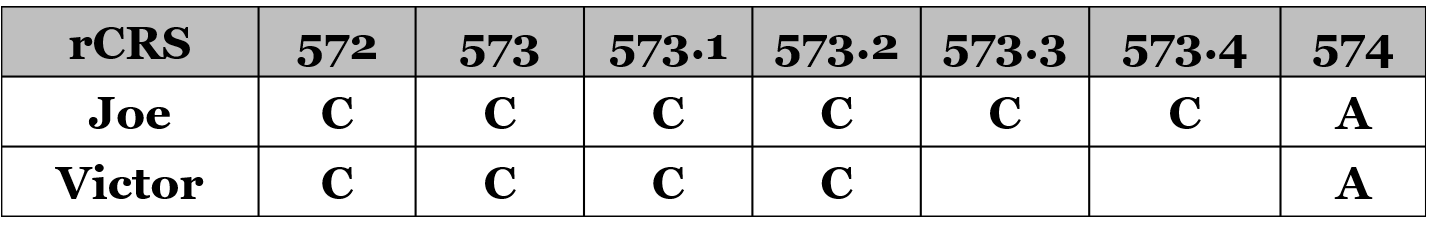
Her results have two additional cytosine bases (“C”) inserted at position 573. I’m reported as having two insertions (573.1C and 573.2C), and she has four insertions ((573.1C, 573.2C, 573.3C, and 573.4C).
This region of the mtDNA has at least 6 cytosine bases (“C”) in a row, from position 568 to 573. In the table below is this stretch of the mtDNA, shown as the revised Cambridge Reference Sequence (rCRS):
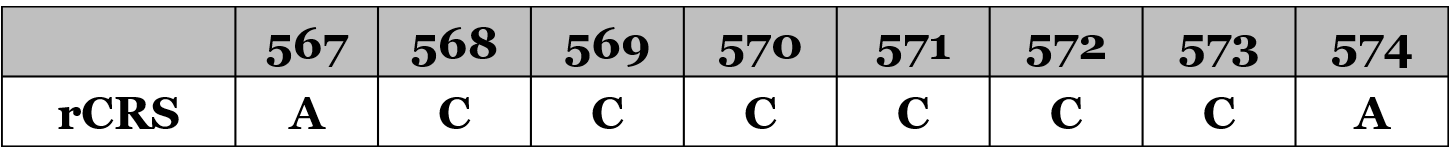
Technically, extra Cs in this region are added to the last C, or 573, out of necessity rather than an actual identification of the insertion location. That’s because there’s no way to determine if a C is inserted between 568 and 569 during replication of the mtDNA genome, or between 569 and 570, so for convenience extra Cs are added at the end of the stretch as 573.1, and so on.
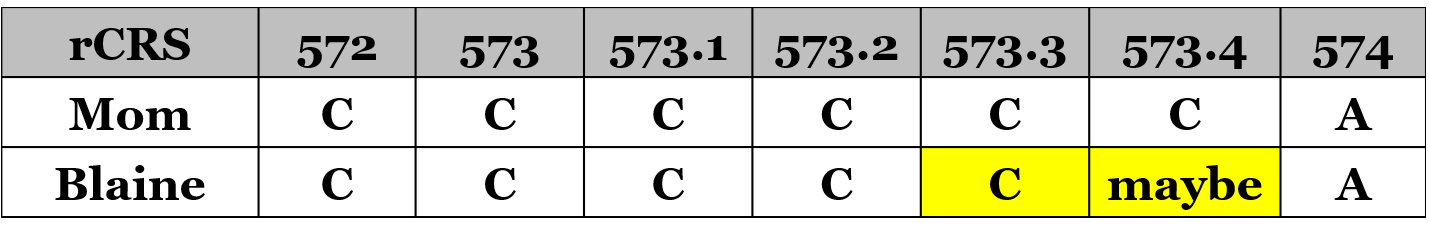
It appears that my mother’s mtDNA has two extra insertions that I don’t have. Perhaps her buccal cells randomly received these two extra Cs during her fetal development while her eggs cell(s) did not have these Cs, or perhaps her egg cell(s) removed two Cs. A comparison of my mom’s poly-cytosine stretch and my stretch look like this:
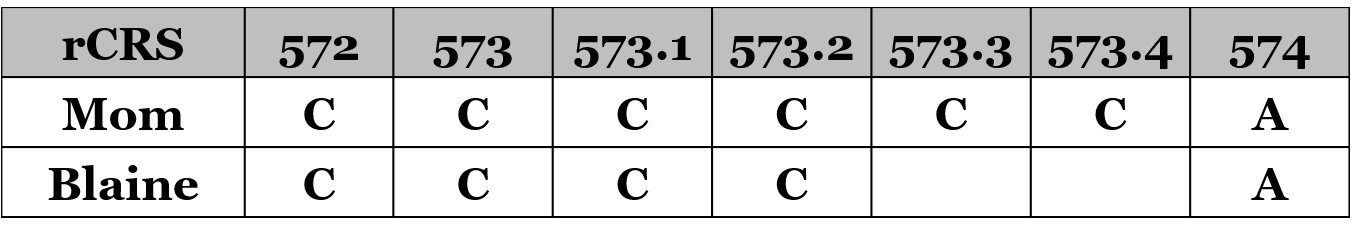
But remember that we’re a genetic distance of four, and this only gets us to two differences (573.3C and 573.4C). Where do the other two differences come from?
Heteroplasmies
We have something like 3 trillion cells in our body, almost all with a copy of our DNA. It shouldn’t be a surprise that there are differences in those three trillion copies. If you have to copy 3 billion letters 3 trillion times, you’d make mistakes too! In reality, we’re a walking bag of millions of different genomes (although the differences are typically only a handful of bases).
The mtDNA genome is no exception. Most people, it is believed, have different versions of mtDNA in their bodies. Having different versions of a genome by the same person is called “heteroplasmy.” Sometimes different cells will have different versions of the mtDNA genome, and sometimes the same cell will have different versions (since a single cell may have tens or hundreds of copies of the mtDNA genome). Once again, these differences are usually a single base pair.
Looking at the first table above, my mother reports 1 heteroplasmy at position 16189. In the rCRS, position 16189 is a “C.” However, my mother reports a “Y”. But Y isn’t one of the four nucleotides, so how can that be?
“Y” is a letter used to represent one of the many different possibilities for mixes of nucleotides at a position. Family Tree DNA provides the heteroplasmy nomenclature:
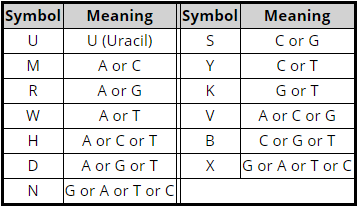
So “Y” means some of my mother’s mtDNA at position 16189 is the wild-type “C” and some of my mother’s mtDNA at position at 16189 is mutated to “T.”
Family Tree DNA can only detect heteroplasmies if the minor allele (meaning the version that is less common in the sample) is present at a concentration of at least 20%. If it is below 20%, the heteroplasmy is too difficult to detect.
It is possible that I have some “T” at position 16189 in some of my mtDNA, although the concentration must be lower than 20%. Heteroplasmies usually take MANY generations to become homoplasmies, to switch from a stable C to a combination of C and T to a stable T. It is extremely unlikely to happen in a single generation. Many and possibly even most of us are in the midst of a heteroplasmy transition. My mtDNA line appears to be in the midst of a transition from C to T at position 16,189.
Adding Evidence – Testing An mtDNA Cousin
I’m lucky to be able to add additional evidence to this analysis. I had my grandmother’s mtDNA first cousin, “Shelly”, autosomal DNA tested at Family Tree DNA, so I upgraded her kit to add an mtDNA test. Like my mother, Shelly inherited her mtDNA from my great-great-grandmother:
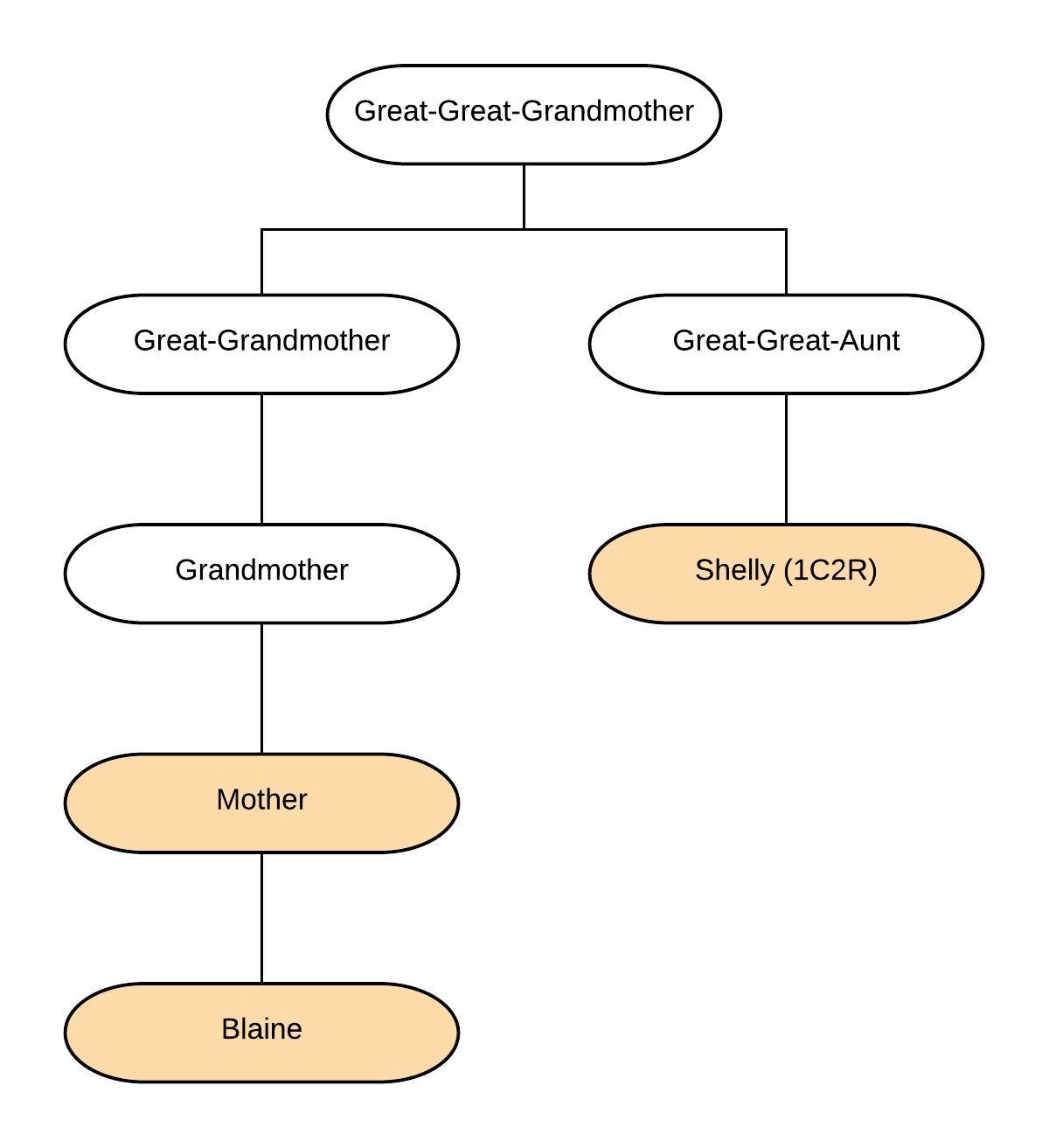
I expected Shelly’s results to resolve the issue of the poly-cytosine stretch around position 573. Was I the anomaly, or was my mother?
Shelly’s results showed that it was me that was the issue! She had the insertions at 573.3C and 573.4C, like my mother. However, unexpected she did not have the heteroplasmy at 16189. That could have gone either way, although my hypothesis was that she would have the heteroplasmy.
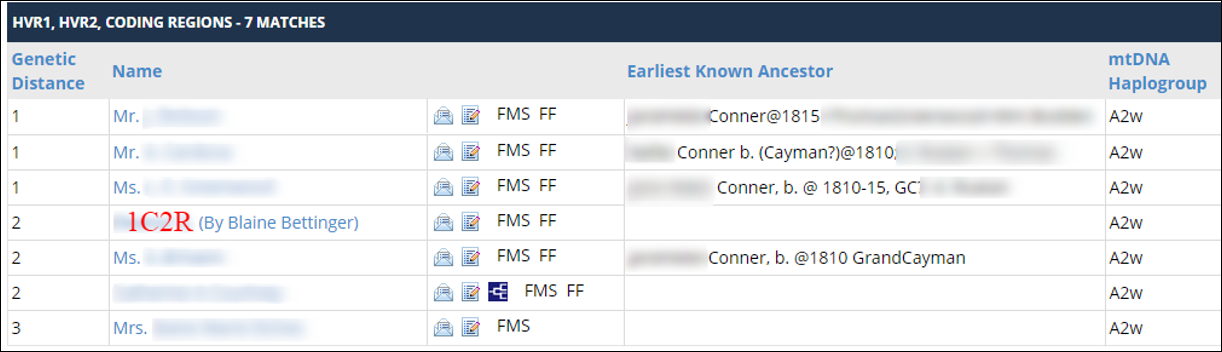
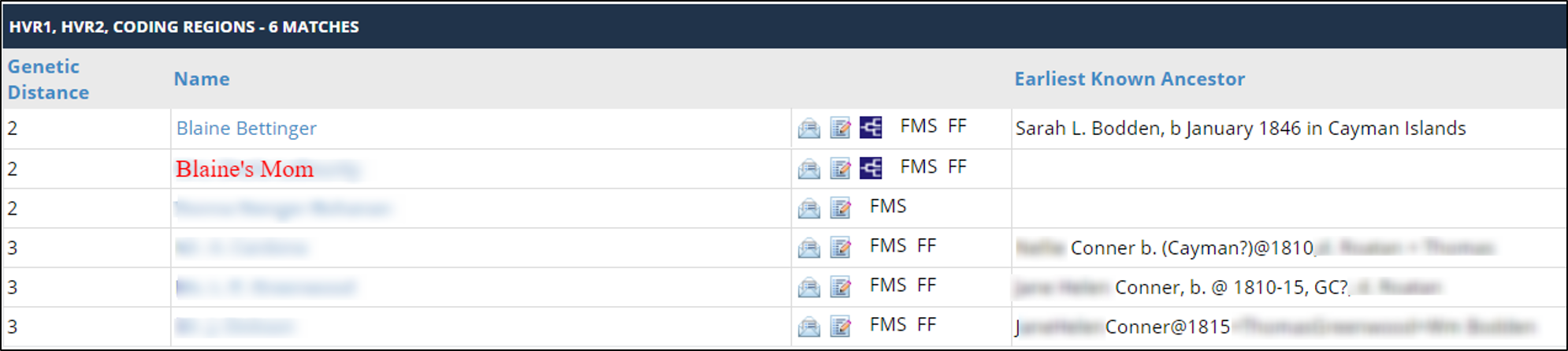
Shelly’s results meant that she matched my mother and I at exactly a genetic distance of 2 each. Here are my mtDNA results, showing that I match Shelly (1C1R) at a distance of 2:
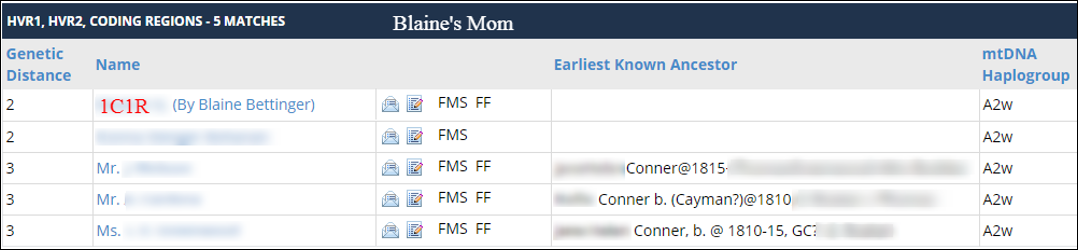
Here are my mother’s results, showing she matches Shelly (1C1R) at a genetic distance of 2:
And here are Shelly’s results, showing that she matches each of us at a genetic distance of 2, acting as a bridge between my mother and me:
Adding Evidence – Additional Testing
I continued to wonder about the extra Cs at position 573. Am I really missing them, or could I possibly have them?
To examine this further, I purchased an mtDNA sequence from YSeq DNA Shop. They offer a variety of mtDNA tests (see their full list of mtDNA tests HERE). There is a full mtDNA sequence for $165 USD, but I really only needed HVR1 (for $25). I thought I would add on HVR2 to also examine the 16189 heteroplasmy. So, for a total of $50, I examined both locations.
Since my DNA was already on file at YSeq, a little less than four weeks later my results were ready:
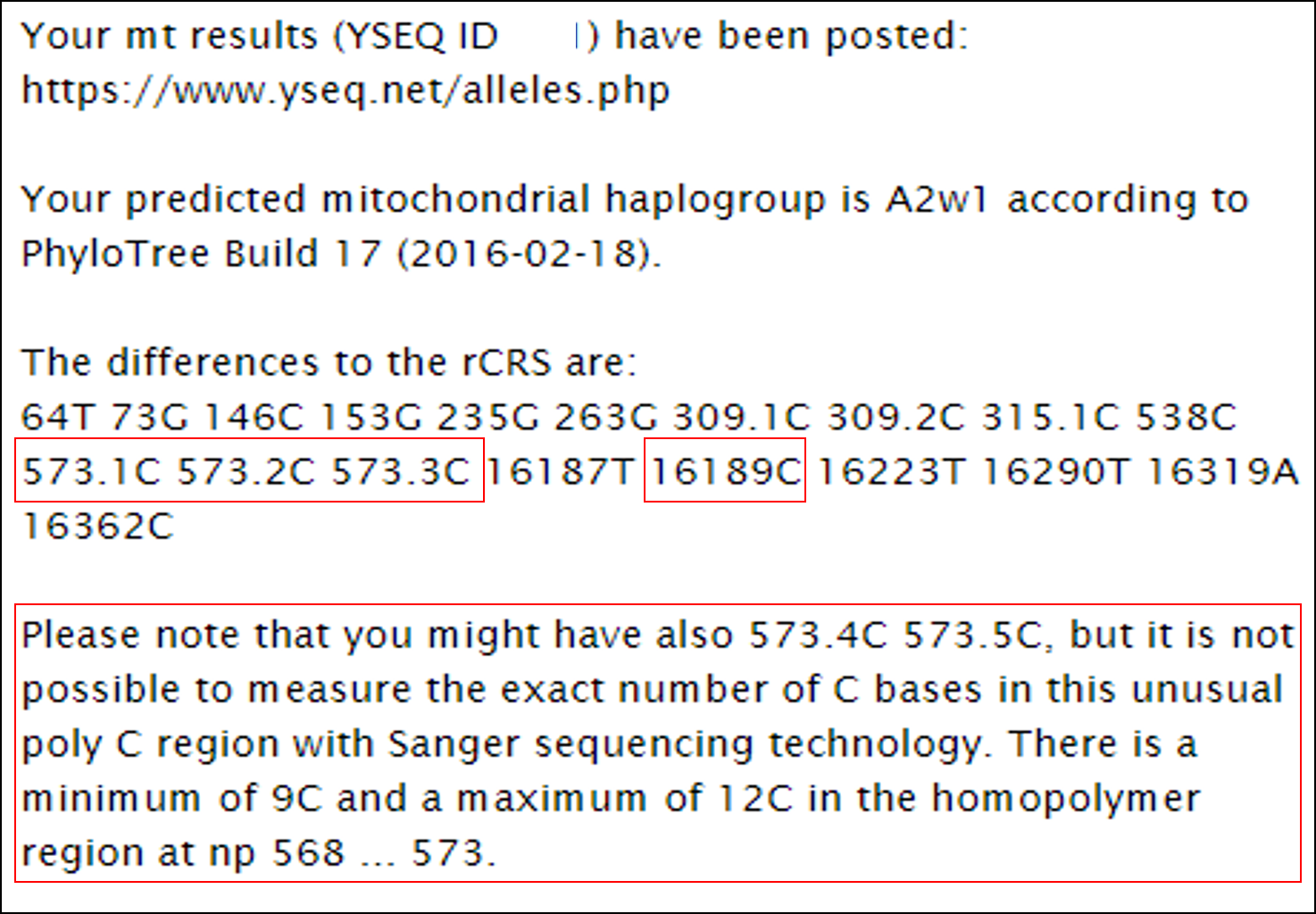
So according to YSeq, I am 16189C (thus not heteroplasmic or not heteroplasmic enough to detect), and I have one extra C at 573.3, and might have 573.4:
Conclusions
This exercise provides insight into my mtDNA line, and why I unexpectedly am not an mtDNA match to my mother despite being a normal atDNA match (yes, she’s my mother!).
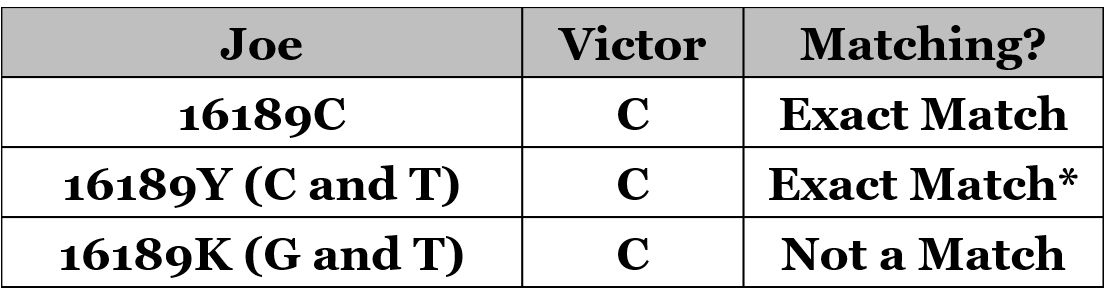
The exercise also suggests that, as others have suggested, heteroplasmies should not be used in the calculation of genetic distance, unless the heteroplasmy reveals that there is no possible match. For example, in the following table, the first two conditions (wild-type and a heteroplasmy showing a C and a T) should show as an exact match between Joe and Victor (GD=0). The third condition, where Joe doesn’t have a “C” at 16189, should correctly be shown as GD=1.
Lastly, the exercise suggests that 573C insertions might potentially be ignored or downweighted when performing comparisons. For example, Joe and Victor might be shown as matches in the following comparison, with an asterisk or other indication of a potential issue in the poly-cytosine stretch at 573:
For More Information
For more about mtDNA, check out the following blog posts:
- The myth of the GD0 – by Judy Russell, another example of heteroplasmy
- DNA Basics: Mitochondrial DNA – by Leah Larkin, an introduction to mtDNA
Thanks for the fuller explanation.
Great info. My mother is from Cayman Brac. We are probably related.
Did you know that the subject of mt-DNA heteroplasmy is not the same in every tissue and this can change over time in a given tissue? I can tell you that you should try to use more resources other than this article (there may be some on this blog if so, let me know) to assimilate this topic.
Thank you for this article, it confirms my own family’s results, that mtDNA is not cast in concrete, and consequently, possibly of little use to an adoptee trying to identify a close match. I FMS tested 11 (with 1 more pending) over 4 generations of a mtDNA line originally from Ireland, and discovered multiple differences between them, including a grandmother who doesn’t show as a match for 2 of her 4 tested grandchildren. The mother of these 2 is an exact match to 1 child, and 2 difference with them, and they do match each other!
mt-DNA heteroplasmy is not the same in every tissue and heteroplasmy may vary over time in a given tissue. Your mother’s ovary may not be reflected in your buccal mucoosa. Thus, in hereditary mt_DNA disorders … which affect energy consuming tissues … one family member may have cardiomyopathy, another renal tubular acidosis or sideroblastic anemia, and another neurological problems — all with the same mt-DNA mutation. Also, identical twins MAY not have identical heteroplasmy and this might be useful in distinguishing twin mothers of maternal cousins.
I’m glad to see this article. Not too long ago, someone who’s a verified descendant of the woman I believe is my second great-grandmother took an mtDNA test with FTDNA. We have the same uncommon haplogroup. We matched on HVR1 and Coding Regions. We didn’t match on HVR2 because she had 3 extra mutations at 573 (same as described in the above article). Our Genetic Distance is 3. I have a great paper trail to prove the relationship and couldn’t understand why the GD wasn’t closer than 3. I didn’t expect “O.” We’re looking back 4 generations to the 1830’s. Our great-grandmothers had different fathers and were raised in different parts of the country. My great-grandmother was adopted at a young age. She had twin daughters, one of whom was my maternal grandmother. There is no way to know if the girls were identical or fraternal. The other twin died at a young age. I think the paper trail is correct.
Interesting topic. I just needed to write an essay on it. But I’m unlikely to be able to fully cope with the essay myself. Therefore, I ordered this essay on the EssayExplorer website – this is a great thing for last minute essay writing – the service is great for any student who has a tight deadline for submitting work.
That sounds like what I’m thinking.
I’m not seeing four differences in the first table. Your mom has two insertions you don’t have and she has one heteroplasmy you don’t have. I assume each one is counted as one mutation so there’s a genetic distance of 3 from those. What is the fourth difference that I’m missing?